Generation of mice deficient in both KLF3/BKLF and KLF8 reveals a genetic interaction and a role for these factors in embryonic globin gene silencing
- PMID: 23716600
- PMCID: PMC3719677
- DOI: 10.1128/MCB.00074-13
Generation of mice deficient in both KLF3/BKLF and KLF8 reveals a genetic interaction and a role for these factors in embryonic globin gene silencing
Abstract
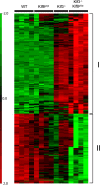
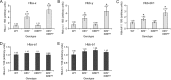
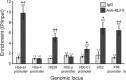
Krüppel-like factors 3 and 8 (KLF3 and KLF8) are highly related transcriptional regulators that bind to similar sequences of DNA. We have previously shown that in erythroid cells there is a regulatory hierarchy within the KLF family, whereby KLF1 drives the expression of both the Klf3 and Klf8 genes and KLF3 in turn represses Klf8 expression. While the erythroid roles of KLF1 and KLF3 have been explored, the contribution of KLF8 to this regulatory network has been unknown. To investigate this, we have generated a mouse model with disrupted KLF8 expression. While these mice are viable, albeit with a reduced life span, mice lacking both KLF3 and KLF8 die at around embryonic day 14.5 (E14.5), indicative of a genetic interaction between these two factors. In the fetal liver, Klf3 Klf8 double mutant embryos exhibit greater dysregulation of gene expression than either of the two single mutants. In particular, we observe derepression of embryonic, but not adult, globin expression. Taken together, these results suggest that KLF3 and KLF8 have overlapping roles in vivo and participate in the silencing of embryonic globin expression during development.
Figures



References
-
- Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. 2008. Kruppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol. 40:1996–2001 - PubMed
-
- Feng WC, Southwood CM, Bieker JJ. 1994. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493–1500 - PubMed
-
- Klevit RE. 1991. Recognition of DNA by Cys2,His2 zinc fingers. Science 253:1367–1393 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
