UGA codon position-dependent incorporation of selenocysteine into mammalian selenoproteins
- PMID: 23716634
- PMCID: PMC3737529
- DOI: 10.1093/nar/gkt409
UGA codon position-dependent incorporation of selenocysteine into mammalian selenoproteins
Abstract
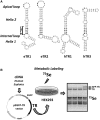
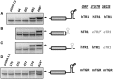
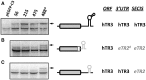
It is thought that the SelenoCysteine Insertion Sequence (SECIS) element and UGA codon are sufficient for selenocysteine (Sec) insertion. However, we found that UGA supported Sec insertion only at its natural position or in its close proximity in mammalian thioredoxin reductase 1 (TR1). In contrast, Sec could be inserted at any tested position in mammalian TR3. Replacement of the 3'-UTR of TR3 with the corresponding segment of a Euplotes crassus TR restricted Sec insertion into the C-terminal region, whereas the 3'-UTR of TR3 conferred unrestricted Sec insertion into E. crassus TR, in which Sec insertion is normally limited to the C-terminal region. Exchanges of 3'-UTRs between mammalian TR1 and E. crassus TR had no effect, as both proteins restricted Sec insertion. We further found that these effects could be explained by the use of selenoprotein-specific SECIS elements. Examination of Sec insertion into other selenoproteins was consistent with this model. The data indicate that mammals evolved the ability to limit Sec insertion into natural positions within selenoproteins, but do so in a selenoprotein-specific manner, and that this process is controlled by the SECIS element in the 3'-UTR.
Figures



References
-
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. - PubMed
-
- Arner ES. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp. Cell Res. 2010;316:1296–1303. - PubMed
-
- Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

