Eosinophils secrete IL-4 to facilitate liver regeneration
- PMID: 23716700
- PMCID: PMC3683773
- DOI: 10.1073/pnas.1304046110
Eosinophils secrete IL-4 to facilitate liver regeneration
Abstract
The liver is a central organ for the synthesis and storage of nutrients, production of serum proteins and hormones, and breakdown of toxins and metabolites. Because the liver is susceptible to toxin- or pathogen-mediated injury, it maintains a remarkable capacity to regenerate by compensatory growth. Specifically, in response to injury, quiescent hepatocytes enter the cell cycle and undergo DNA replication to promote liver regrowth. Despite the elucidation of a number of regenerative factors, the mechanisms by which liver injury triggers hepatocyte proliferation are incompletely understood. We demonstrate here that eosinophils stimulate liver regeneration after partial hepatectomy and toxin-mediated injury. Liver injury results in rapid recruitment of eosinophils, which secrete IL-4 to promote the proliferation of quiescent hepatocytes. Surprisingly, signaling via the IL-4Rα in macrophages, which have been implicated in tissue repair, is dispensable for hepatocyte proliferation and liver regrowth after injury. Instead, IL-4 exerts its proliferative actions via IL-4Rα in hepatocytes. Our findings thus provide a unique mechanism by which eosinophil-derived IL-4 stimulates hepatocyte proliferation in regenerating liver.
Keywords: inflammation; parasites; tissue injury and repair; type 2 immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

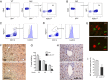
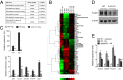
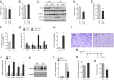
References
-
- Taub R. Liver regeneration: From myth to mechanism. Nat Rev Mol Cell Biol. 2004;5(10):836–847. - PubMed
-
- Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol (Chic) 1931;12:186–202.
-
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276(5309):60–66. - PubMed
-
- Huang W, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312(5771):233–236. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

