Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications
- PMID: 23717205
- PMCID: PMC3662640
- DOI: 10.1371/journal.ppat.1003375
Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications
Abstract
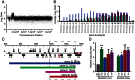
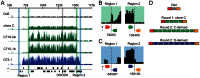
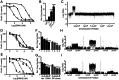
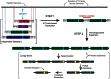
Malaria drug resistance contributes to up to a million annual deaths. Judicious deployment of new antimalarials and vaccines could benefit from an understanding of early molecular events that promote the evolution of parasites. Continuous in vitro challenge of Plasmodium falciparum parasites with a novel dihydroorotate dehydrogenase (DHODH) inhibitor reproducibly selected for resistant parasites. Genome-wide analysis of independently-derived resistant clones revealed a two-step strategy to evolutionary success. Some haploid blood-stage parasites first survive antimalarial pressure through fortuitous DNA duplications that always included the DHODH gene. Independently-selected parasites had different sized amplification units but they were always flanked by distant A/T tracks. Higher level amplification and resistance was attained using a second, more efficient and more accurate, mechanism for head-to-tail expansion of the founder unit. This second homology-based process could faithfully tune DNA copy numbers in either direction, always retaining the unique DNA amplification sequence from the original A/T-mediated duplication for that parasite line. Pseudo-polyploidy at relevant genomic loci sets the stage for gaining additional mutations at the locus of interest. Overall, we reveal a population-based genomic strategy for mutagenesis that operates in human stages of P. falciparum to efficiently yield resistance-causing genetic changes at the correct locus in a successful parasite. Importantly, these founding events arise with precision; no other new amplifications are seen in the resistant haploid blood stage parasite. This minimizes the need for meiotic genetic cleansing that can only occur in sexual stage development of the parasite in mosquitoes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- White NJ (1992) Antimalarial drug resistance: the pace quickens. J Antimicrob Chemother 30: 571–585. - PubMed
-
- Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, et al. (2012) Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379: 413–431. - PubMed
-
- WHO (2012) World Malaria Report.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

