Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia
- PMID: 23717306
- PMCID: PMC3651956
- DOI: 10.3389/fmicb.2013.00115
Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia
Abstract
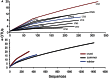
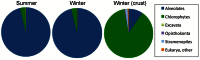
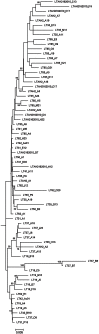
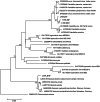
This study describes the community structure of the microbial eukaryotic community from hypersaline Lake Tyrrell, Australia, using near full length 18S rRNA sequences. Water samples were taken in both summer and winter over a 4-year period. The extent of eukaryotic diversity detected was low, with only 35 unique phylotypes using a 97% sequence similarity threshold. The water samples were dominated (91%) by a novel cluster of the Alveolate, Apicomplexa Colpodella spp., most closely related to C. edax. The Chlorophyte, Dunaliella spp. accounted for less than 35% of water column samples. However, the eukaryotic community entrained in a salt crust sample was vastly different and was dominated (83%) by the Dunaliella spp. The patterns described here represent the first observation of microbial eukaryotic dynamics in this system and provide a multiyear comparison of community composition by season. The lack of expected seasonal distribution in eukaryotic communities paired with abundant nanoflagellates suggests that grazing may significantly structure microbial eukaryotic communities in this system.
Keywords: 18S rRNA; Colpodella; Dunaliella; diversity; hypersaline; microbial eukaryotes; saltern.
Figures





Similar articles
-
Seasonal dynamics of eukaryotic microbial diversity in hypersaline Tuz Lake characterized by 18S rDNA sequencing.J Eukaryot Microbiol. 2023 Nov-Dec;70(6):e12993. doi: 10.1111/jeu.12993. Epub 2023 Aug 1. J Eukaryot Microbiol. 2023. PMID: 37528557
-
Eukaryotic microbial communities in hypersaline soils and sediments from the alkaline hypersaline Huama Lake as revealed by 454 pyrosequencing.Antonie Van Leeuwenhoek. 2014 May;105(5):871-80. doi: 10.1007/s10482-014-0141-4. Epub 2014 Feb 22. Antonie Van Leeuwenhoek. 2014. PMID: 24563154
-
Microbial Diversity of Hypersaline Sediments from Lake Lucero Playa in White Sands National Monument, New Mexico, USA.Microb Ecol. 2018 Aug;76(2):404-418. doi: 10.1007/s00248-018-1142-z. Epub 2018 Jan 29. Microb Ecol. 2018. PMID: 29380029
-
Distribution and diversity of eukaryotic microalgae in Kuwait waters assessed using 18S rRNA gene sequencing.PLoS One. 2021 Apr 26;16(4):e0250645. doi: 10.1371/journal.pone.0250645. eCollection 2021. PLoS One. 2021. PMID: 33901235 Free PMC article.
-
Strong Seasonality in Arctic Estuarine Microbial Food Webs.Front Microbiol. 2019 Nov 29;10:2628. doi: 10.3389/fmicb.2019.02628. eCollection 2019. Front Microbiol. 2019. PMID: 31849850 Free PMC article. Review.
Cited by
-
Microbial diversity in polyextreme salt flats and their potential applications.Environ Sci Pollut Res Int. 2024 Feb;31(8):11371-11405. doi: 10.1007/s11356-023-31644-9. Epub 2024 Jan 5. Environ Sci Pollut Res Int. 2024. PMID: 38180652 Review.
-
A Review on Viral Metagenomics in Extreme Environments.Front Microbiol. 2019 Oct 18;10:2403. doi: 10.3389/fmicb.2019.02403. eCollection 2019. Front Microbiol. 2019. PMID: 31749771 Free PMC article. Review.
-
High genetic diversity and novelty in eukaryotic plankton assemblages inhabiting saline lakes in the Qaidam basin.PLoS One. 2014 Nov 17;9(11):e112812. doi: 10.1371/journal.pone.0112812. eCollection 2014. PLoS One. 2014. PMID: 25401703 Free PMC article.
-
Comparative study of multiple approaches for identifying cultivable microalgae population diversity from freshwater samples.PLoS One. 2023 Jul 7;18(7):e0285913. doi: 10.1371/journal.pone.0285913. eCollection 2023. PLoS One. 2023. PMID: 37418475 Free PMC article.
-
The ecology of Dunaliella in high-salt environments.J Biol Res (Thessalon). 2014 Dec 18;21(1):23. doi: 10.1186/s40709-014-0023-y. eCollection 2014 Dec. J Biol Res (Thessalon). 2014. PMID: 25984505 Free PMC article. Review.
References
-
- Avron M., Ben-Amotz A. (1979). Metabolic adaptation of the alga Dunaliella to low water activity, in Strategies of Microbial Life in Extreme Environments, ed Shilo M. (Berlin: Dahlem Konferenzen; ), 83–91
-
- Ben-Amotz A., Polle J. E. W., Rao D. V. S. (2009). The Alga Dunaliella: Biodiversity, Physiology, Genomics, and Biotechnology. Enfield, NH: Science Publishers
-
- Borowitzka M. (2007). The taxonomy of the genus Dunaliella (Chlorophyta, Dunaliellales) with emphasis on the marine and halophilic species. J. Appl. Phycol. 19, 567–590
-
- Borowitzka M. A. (1990). The mass culture of Dunaliella salina, in Technical Resource Papers Regional Workshop on the Culture and Utilization of Seaweads. Vol. 2 Fisheries and Aquaculture Department. FAO Corporate Document Repository. Available online at: http://www.fao.org/docrep/field/003/AB728E/AB728E00.htm
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

