Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae
- PMID: 23717440
- PMCID: PMC3661542
- DOI: 10.1371/journal.pone.0063512
Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae
Abstract
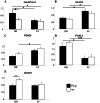
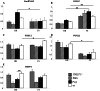
Despite massive research efforts, the molecular etiology of bovine polledness and the developmental pathways involved in horn ontogenesis are still poorly understood. In a recent article, we provided evidence for the existence of at least two different alleles at the Polled locus and identified candidate mutations for each of them. None of these mutations was located in known coding or regulatory regions, thus adding to the complexity of understanding the molecular basis of polledness. We confirm previous results here and exhaustively identify the causative mutation for the Celtic allele (PC) and four candidate mutations for the Friesian allele (PF). We describe a previously unreported eyelash-and-eyelid phenotype associated with regular polledness, and present unique histological and gene expression data on bovine horn bud differentiation in fetuses affected by three different horn defect syndromes, as well as in wild-type controls. We propose the ectopic expression of a lincRNA in PC/p horn buds as a probable cause of horn bud agenesis. In addition, we provide evidence for an involvement of OLIG2, FOXL2 and RXFP2 in horn bud differentiation, and draw a first link between bovine, ovine and caprine Polled loci. Our results represent a first and important step in understanding the genetic pathways and key process involved in horn bud differentiation in Bovidae.
Conflict of interest statement
Figures



References
-
- Prayaga KC (2007) Genetic options to replace dehorning in beef cattle-a review. Aust J Agric Res 58: 1–8.
-
- Graf B, Senn M (1999) Behavioural and physiological responses of calves to dehorning by heat cauterization with or without local anaesthesia. Applied Animal Behaviour Science 62: 153–171.
-
- Roman A (2004) L’élevage bovin en Egypte antique. Bull Soc Fr Hist Méd Sci Vét 3: 35–45.
-
- Capitan A (2010) Détection et caractérisation de loci responsables d’anomalies de cornage en race Charolais français [PhD thesis]. Paris: Institut des Sciences et Industries du Vivant et de l’Environnement (Agro Paris Tech).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

