Comprehensive functional annotation of seventy-one breast cancer risk Loci
- PMID: 23717510
- PMCID: PMC3661550
- DOI: 10.1371/journal.pone.0063925
Comprehensive functional annotation of seventy-one breast cancer risk Loci
Abstract
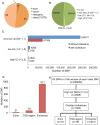
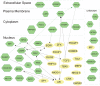
Breast Cancer (BCa) genome-wide association studies revealed allelic frequency differences between cases and controls at index single nucleotide polymorphisms (SNPs). To date, 71 loci have thus been identified and replicated. More than 320,000 SNPs at these loci define BCa risk due to linkage disequilibrium (LD). We propose that BCa risk resides in a subgroup of SNPs that functionally affects breast biology. Such a shortlist will aid in framing hypotheses to prioritize a manageable number of likely disease-causing SNPs. We extracted all the SNPs, residing in 1 Mb windows around breast cancer risk index SNP from the 1000 genomes project to find correlated SNPs. We used FunciSNP, an R/Bioconductor package developed in-house, to identify potentially functional SNPs at 71 risk loci by coinciding them with chromatin biofeatures. We identified 1,005 SNPs in LD with the index SNPs (r(2)≥0.5) in three categories; 21 in exons of 18 genes, 76 in transcription start site (TSS) regions of 25 genes, and 921 in enhancers. Thirteen SNPs were found in more than one category. We found two correlated and predicted non-benign coding variants (rs8100241 in exon 2 and rs8108174 in exon 3) of the gene, ANKLE1. Most putative functional LD SNPs, however, were found in either epigenetically defined enhancers or in gene TSS regions. Fifty-five percent of these non-coding SNPs are likely functional, since they affect response element (RE) sequences of transcription factors. Functionality of these SNPs was assessed by expression quantitative trait loci (eQTL) analysis and allele-specific enhancer assays. Unbiased analyses of SNPs at BCa risk loci revealed new and overlooked mechanisms that may affect risk of the disease, thereby providing a valuable resource for follow-up studies.
Conflict of interest statement
Figures




References
-
- Peng S, Lu B, Ruan W, Zhu Y, Sheng H, et al. (2011) Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast cancer research and treatment 127: 309–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

