IMSA: integrated metagenomic sequence analysis for identification of exogenous reads in a host genomic background
- PMID: 23717627
- PMCID: PMC3662787
- DOI: 10.1371/journal.pone.0064546
IMSA: integrated metagenomic sequence analysis for identification of exogenous reads in a host genomic background
Abstract
Metagenomics, the study of microbial genomes within diverse environments, is a rapidly developing field. The identification of microbial sequences within a host organism enables the study of human intestinal, respiratory, and skin microbiota, and has allowed the identification of novel viruses in diseases such as Merkel cell carcinoma. There are few publicly available tools for metagenomic high throughput sequence analysis. We present Integrated Metagenomic Sequence Analysis (IMSA), a flexible, fast, and robust computational analysis pipeline that is available for public use. IMSA takes input sequence from high throughput datasets and uses a user-defined host database to filter out host sequence. IMSA then aligns the filtered reads to a user-defined universal database to characterize exogenous reads within the host background. IMSA assigns a score to each node of the taxonomy based on read frequency, and can output this as a taxonomy report suitable for cluster analysis or as a taxonomy map (TaxMap). IMSA also outputs the specific sequence reads assigned to a taxon of interest for downstream analysis. We demonstrate the use of IMSA to detect pathogens and normal flora within sequence data from a primary human cervical cancer carrying HPV16, a primary human cutaneous squamous cell carcinoma carrying HPV 16, the CaSki cell line carrying HPV16, and the HeLa cell line carrying HPV18.
Conflict of interest statement
Figures

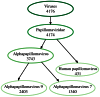

References
-
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5: R245–249. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

