Integrated analysis of recurrent properties of cancer genes to identify novel drivers
- PMID: 23718799
- PMCID: PMC4054099
- DOI: 10.1186/gb-2013-14-5-r52
Integrated analysis of recurrent properties of cancer genes to identify novel drivers
Abstract
The heterogeneity of cancer genomes in terms of acquired mutations complicates the identification of genes whose modification may exert a driver role in tumorigenesis. In this study, we present a novel method that integrates expression profiles, mutation effects, and systemic properties of mutated genes to identify novel cancer drivers. We applied our method to ovarian cancer samples and were able to identify putative drivers in the majority of carcinomas without mutations in known cancer genes, thus suggesting that it can be used as a complementary approach to find rare driver mutations that cannot be detected using frequency-based approaches.
Figures



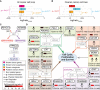
References
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D. et al.Patterns of somatic mutation in human cancer genomes. Nature. 2007;14:153–158. doi: 10.1038/nature05610. - DOI - PMC - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV. et al.The genomic landscapes of human breast and colorectal cancers. Science. 2007;14:1108–1113. doi: 10.1126/science.1145720. - DOI - PubMed
-
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K. et al.Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;14:1069–1075. doi: 10.1038/nature07423. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

