Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions
- PMID: 23720318
- PMCID: PMC3690903
- DOI: 10.1073/pnas.1221210110
Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions
Abstract
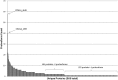
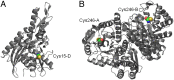
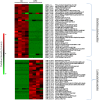
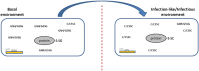
Characterization of the mature protein complement in cells is crucial for a better understanding of cellular processes on a systems-wide scale. Toward this end, we used single-dimension ultra-high-pressure liquid chromatography mass spectrometry to investigate the comprehensive "intact" proteome of the Gram-negative bacterial pathogen Salmonella Typhimurium. Top-down proteomics analysis revealed 563 unique proteins including 1,665 proteoforms generated by posttranslational modifications (PTMs), representing the largest microbial top-down dataset reported to date. We confirmed many previously recognized aspects of Salmonella biology and bacterial PTMs, and our analysis also revealed several additional biological insights. Of particular interest was differential utilization of the protein S-thiolation forms S-glutathionylation and S-cysteinylation in response to infection-like conditions versus basal conditions. This finding of a S-glutathionylation-to-S-cysteinylation switch in a condition-specific manner was corroborated by bottom-up proteomics data and further by changes in corresponding biosynthetic pathways under infection-like conditions and during actual infection of host cells. This differential utilization highlights underlying metabolic mechanisms that modulate changes in cellular signaling, and represents a report of S-cysteinylation in Gram-negative bacteria. Additionally, the functional relevance of these PTMs was supported by protein structure and gene deletion analyses. The demonstrated utility of our simple proteome-wide intact protein level measurement strategy for gaining biological insight should promote broader adoption and applications of top-down proteomics approaches.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, Stephenson JL., Jr Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal Chem. 2008;80(5):1459–1467. - PubMed
-
- Tsai YS, et al. Precursor ion independent algorithm for top-down shotgun proteomics. J Am Soc Mass Spectrom. 2009;20(11):2154–2166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

