Phylogenetic and Functional Analysis of Metagenome Sequence from High-Temperature Archaeal Habitats Demonstrate Linkages between Metabolic Potential and Geochemistry
- PMID: 23720654
- PMCID: PMC3654217
- DOI: 10.3389/fmicb.2013.00095
Phylogenetic and Functional Analysis of Metagenome Sequence from High-Temperature Archaeal Habitats Demonstrate Linkages between Metabolic Potential and Geochemistry
Abstract
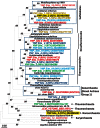
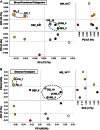
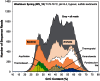
Geothermal habitats in Yellowstone National Park (YNP) provide an unparalleled opportunity to understand the environmental factors that control the distribution of archaea in thermal habitats. Here we describe, analyze, and synthesize metagenomic and geochemical data collected from seven high-temperature sites that contain microbial communities dominated by archaea relative to bacteria. The specific objectives of the study were to use metagenome sequencing to determine the structure and functional capacity of thermophilic archaeal-dominated microbial communities across a pH range from 2.5 to 6.4 and to discuss specific examples where the metabolic potential correlated with measured environmental parameters and geochemical processes occurring in situ. Random shotgun metagenome sequence (∼40-45 Mb Sanger sequencing per site) was obtained from environmental DNA extracted from high-temperature sediments and/or microbial mats and subjected to numerous phylogenetic and functional analyses. Analysis of individual sequences (e.g., MEGAN and G + C content) and assemblies from each habitat type revealed the presence of dominant archaeal populations in all environments, 10 of whose genomes were largely reconstructed from the sequence data. Analysis of protein family occurrence, particularly of those involved in energy conservation, electron transport, and autotrophic metabolism, revealed significant differences in metabolic strategies across sites consistent with differences in major geochemical attributes (e.g., sulfide, oxygen, pH). These observations provide an ecological basis for understanding the distribution of indigenous archaeal lineages across high-temperature systems of YNP.
Keywords: archaea; functional genomics; geochemistry; phylogeny; thermophilic archaea and bacteria.
Figures







References
-
- Auernik K. S., Kelly R. M. (2008). Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74, 7723–773210.1128/AEM.01545-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

