Three infectious viral species lying in wait in the banana genome
- PMID: 23720724
- PMCID: PMC3719817
- DOI: 10.1128/JVI.00899-13
Three infectious viral species lying in wait in the banana genome
Abstract
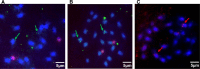
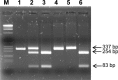
Plant pararetroviruses integrate serendipitously into their host genomes. The banana genome harbors integrated copies of banana streak virus (BSV) named endogenous BSV (eBSV) that are able to release infectious pararetrovirus. In this investigation, we characterized integrants of three BSV species-Goldfinger (eBSGFV), Imove (eBSImV), and Obino l'Ewai (eBSOLV)-in the seedy Musa balbisiana Pisang klutuk wulung (PKW) by studying their molecular structure, genomic organization, genomic landscape, and infectious capacity. All eBSVs exhibit extensive viral genome duplications and rearrangements. eBSV segregation analysis on an F1 population of PKW combined with fluorescent in situ hybridization analysis showed that eBSImV, eBSOLV, and eBSGFV are each present at a single locus. eBSOLV and eBSGFV contain two distinct alleles, whereas eBSImV has two structurally identical alleles. Genotyping of both eBSV and viral particles expressed in the progeny demonstrated that only one allele for each species is infectious. The infectious allele of eBSImV could not be identified since the two alleles are identical. Finally, we demonstrate that eBSGFV and eBSOLV are located on chromosome 1 and eBSImV is located on chromosome 2 of the reference Musa genome published recently. The structure and evolution of eBSVs suggest sequential integration into the plant genome, and haplotype divergence analysis confirms that the three loci display differential evolution. Based on our data, we propose a model for BSV integration and eBSV evolution in the Musa balbisiana genome. The mutual benefits of this unique host-pathogen association are also discussed.
Figures


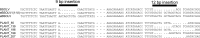
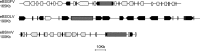


References
-
- Vogt PK. 1997. Historical introduction to the general properties of retroviruses, p 1–26 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

