Comparing and combining capillary electrophoresis electrospray ionization mass spectrometry and nano-liquid chromatography electrospray ionization mass spectrometry for the characterization of post-translationally modified histones
- PMID: 23720761
- PMCID: PMC3769336
- DOI: 10.1074/mcp.M112.024109
Comparing and combining capillary electrophoresis electrospray ionization mass spectrometry and nano-liquid chromatography electrospray ionization mass spectrometry for the characterization of post-translationally modified histones
Abstract
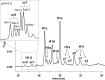
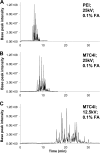
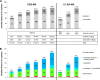
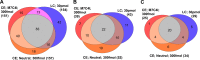
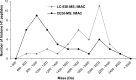
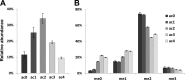
We present the first comprehensive capillary electrophoresis electrospray ionization mass spectrometry (CESI-MS) analysis of post-translational modifications derived from H1 and core histones. Using a capillary electrophoresis system equipped with a sheathless high-sensitivity porous sprayer and nano-liquid chromatography electrospray ionization mass spectrometry (nano-LC-ESI-MS) as two complementary techniques, we characterized H1 histones isolated from rat testis. Without any pre-separation of the perchloric acid extraction, a total of 70 different modified peptides, including 50 phosphopeptides, were identified in the rat linker histones H1.0, H1a-H1e, and H1t. Out of the 70 modified H1 histone peptides, 27 peptides could be identified with CESI-MS only, and 11 solely with LC-ESI-MS. Immobilized metal-affinity chromatography enrichment prior to MS analysis yielded a total of 55 phosphopeptides; 22 of these peptides could be identified only by CESI-MS, and 19 only by LC-ESI-MS, showing the complementarity of the two techniques. We mapped 42 H1 modification sites, including 31 phosphorylation sites, of which 8 were novel sites. For the analysis of core histones, we chose a different strategy. In a first step, the sulfuric-acid-extracted core histones were pre-separated using reverse-phase high-performance liquid chromatography. Individual rat testis core histone fractions obtained in this way were digested and analyzed via bottom-up CESI-MS. This approach yielded the identification of 42 different modification sites including acetylation (lysine and N(α)-terminal); mono-, di-, and trimethylation; and phosphorylation. When we applied CESI-MS for the analysis of intact core histone subtypes from butyrate-treated mouse tumor cells, we were able to rapidly detect their degree of modification, and we found this method very useful for the separation of isobaric trimethyl and acetyl modifications. Taken together, our results highlight the need for additional techniques for the comprehensive analysis of post-translational modifications. CESI-MS is a promising new proteomics tool as demonstrated by this, the first comprehensive analysis of histone modifications, using rat testis as an example.
Figures



References
-
- Izzo A., Kamieniarz K., Schneider R. (2008) The histone H1 family: specific members, specific functions? Biol. Chem. 389, 333–343 - PubMed
-
- Happel N., Doenecke D. (2009) Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431, 1–12 - PubMed
-
- Wood C., Snijders A., Williamson J., Reynolds C., Baldwin J., Dickman M. (2009) Post-translational modifications of the linker histone variants and their association with cell mechanisms. FEBS J. 276, 3685–3697 - PubMed
-
- Turner B. M. (2000) Histone acetylation and an epigenetic code. Bioessays 22, 836–845 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

