Complete TCR-α gene locus control region activity in T cells derived in vitro from embryonic stem cells
- PMID: 23720809
- PMCID: PMC3755507
- DOI: 10.4049/jimmunol.1300521
Complete TCR-α gene locus control region activity in T cells derived in vitro from embryonic stem cells
Abstract
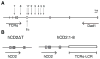
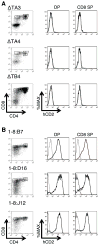
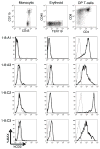
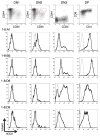
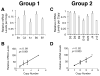
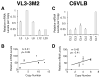
Locus control regions (LCRs) are cis-acting gene regulatory elements with the unique, integration site-independent ability to transfer the characteristics of their locus-of-origin's gene expression pattern to a linked transgene in mice. LCR activities have been discovered in numerous T cell lineage-expressed gene loci. These elements can be adapted to the design of stem cell gene therapy vectors that direct robust therapeutic gene expression to the T cell progeny of engineered stem cells. Currently, transgenic mice provide the only experimental approach that wholly supports all the critical aspects of LCR activity. In this study, we report the manifestation of all key features of mouse TCR-α gene LCR function in T cells derived in vitro from mouse embryonic stem cells. High-level, copy number-related TCR-α LCR-linked reporter gene expression levels are cell type restricted in this system, and upregulated during the expected stage transition of T cell development. We also report that de novo introduction of TCR-α LCR-linked transgenes into existing T cell lines yields incomplete LCR activity. These data indicate that establishing full TCR-α LCR activity requires critical molecular events occurring prior to final T lineage determination. This study also validates a novel, tractable, and more rapid approach for the study of LCR activity in T cells, and its translation to therapeutic genetic engineering.
Figures
References
-
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. - PubMed
-
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. - PubMed
-
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

