Amino acid sequence diversity of the major human papillomavirus capsid protein: implications for current and next generation vaccines
- PMID: 23722024
- PMCID: PMC3769806
- DOI: 10.1016/j.meegid.2013.05.013
Amino acid sequence diversity of the major human papillomavirus capsid protein: implications for current and next generation vaccines
Abstract
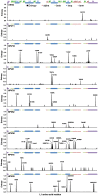
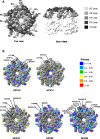
Despite the fidelity of host cell polymerases, the human papillomavirus (HPV) displays a degree of genomic polymorphism resulting in distinct genotypes and intra-type variants. The current HPV vaccines target the most prevalent genotypes associated with cervical cancer (HPV16/18) and genital warts (HPV6/11). Although these vaccines confer some measure of cross-protection, a multivalent HPV vaccine is in the pipeline that aims to broaden vaccine protection against other cervical cancer-associated genotypes including HPV31, HPV33, HPV45, HPV52 and HPV58. Both current and next generation vaccines comprise virus-like particles, based upon the major capsid protein, L1, and vaccine-induced, type-specific protection is likely mediated by neutralizing antibodies targeting L1 surface-exposed domains. The aim of this study was to perform an in silico analysis of existing full length L1 sequences representing vaccine-relevant HPV genotypes in order to address the degree of naturally-occurring, intra-type polymorphisms. In total, 1281 sequences from the Americas, Africa, Asia and Europe were assembled. Intra-type entropy was low and/or limited to non-surface-exposed residues for HPV6, HPV11 and HPV52 suggesting a minimal effect on vaccine antibodies for these genotypes. For HPV16, intra-type entropy was high but the present analysis did not reveal any significant polymorphisms not previously identified. For HPV31, HPV33, HPV58, however, intra-type entropy was high, mostly mapped to surface-exposed domains and in some cases within known neutralizing antibody epitopes. For HPV18 and HPV45 there were too few sequences for a definitive analysis, but HPV45 displayed some degree of surface-exposed residue diversity. In most cases, the reference sequence for each genotype represented a minority variant and the consensus L1 sequences for HPV18, HPV31, HPV45 and HPV58 did not reflect the L1 sequence of the currently available HPV pseudoviruses. These data highlight a number of variant amino acid residues that warrant further investigation for vaccine and natural history studies of HPV.
Keywords: Diversity; Entropy; HPV; L1; Papillomavirus.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Comprehensive Assessment of the Antigenic Impact of Human Papillomavirus Lineage Variation on Recognition by Neutralizing Monoclonal Antibodies Raised against Lineage A Major Capsid Proteins of Vaccine-Related Genotypes.J Virol. 2020 Nov 23;94(24):e01236-20. doi: 10.1128/JVI.01236-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967963 Free PMC article.
-
Worldwide genetic variations in high-risk human papillomaviruses capsid L1 gene and their impact on vaccine efficiency.Gene. 2021 May 25;782:145533. doi: 10.1016/j.gene.2021.145533. Epub 2021 Feb 23. Gene. 2021. PMID: 33636291 Review.
-
Pre-clinical immunogenicity of human papillomavirus alpha-7 and alpha-9 major capsid proteins.Vaccine. 2014 Nov 12;32(48):6548-55. doi: 10.1016/j.vaccine.2014.07.116. Epub 2014 Sep 6. Vaccine. 2014. PMID: 25203446 Free PMC article.
-
Sensitivity of Human Papillomavirus (HPV) Lineage and Sublineage Variant Pseudoviruses to Neutralization by Nonavalent Vaccine Antibodies.J Infect Dis. 2019 Nov 6;220(12):1940-1945. doi: 10.1093/infdis/jiz401. J Infect Dis. 2019. PMID: 31412122 Free PMC article.
-
Developments in L2-based human papillomavirus (HPV) vaccines.Virus Res. 2017 Mar 2;231:166-175. doi: 10.1016/j.virusres.2016.11.020. Epub 2016 Nov 23. Virus Res. 2017. PMID: 27889616 Free PMC article. Review.
Cited by
-
Rational design of a multi-valent human papillomavirus vaccine by capsomere-hybrid co-assembly of virus-like particles.Nat Commun. 2020 Jun 5;11(1):2841. doi: 10.1038/s41467-020-16639-1. Nat Commun. 2020. PMID: 32503989 Free PMC article.
-
Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.mBio. 2017 Sep 26;8(5):e00787-17. doi: 10.1128/mBio.00787-17. mBio. 2017. PMID: 28951471 Free PMC article.
-
Naturally Occurring Single Amino Acid Substitution in the L1 Major Capsid Protein of Human Papillomavirus Type 16: Alteration of Susceptibility to Antibody-Mediated Neutralization.J Infect Dis. 2017 Oct 17;216(7):867-876. doi: 10.1093/infdis/jix274. J Infect Dis. 2017. PMID: 28968823 Free PMC article.
-
Lack of detectable HPV18 antibodies in 14% of quadrivalent vaccinees in a longitudinal cohort study.NPJ Vaccines. 2024 Aug 13;9(1):146. doi: 10.1038/s41541-024-00941-w. NPJ Vaccines. 2024. PMID: 39138224 Free PMC article.
-
The DE and FG loops of the HPV major capsid protein contribute to the epitopes of vaccine-induced cross-neutralising antibodies.Sci Rep. 2016 Dec 22;6:39730. doi: 10.1038/srep39730. Sci Rep. 2016. PMID: 28004837 Free PMC article. Clinical Trial.
References
-
- Arbyn M., Castellsague X., de Sanjose S., Bruni L., Saraiya M., Bray F., Ferlay J. Worldwide burden of cervical cancer in 2008. Ann. Oncol. 2011;22:2675–2686. - PubMed
-
- Bernard H.U., Calleja-Macias I.E., Dunn S.T. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int. J. Cancer. 2006;118:1071–1076. - PubMed
-
- Bishop B., Dasgupta J., Klein M., Garcea R.L., Christensen N.D., Zhao R., Chen X.S. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J. Biol. Chem. 2007;282:31803–31811. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

