A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C
- PMID: 23723239
- PMCID: PMC3763809
- DOI: 10.1126/science.1235532
A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C
Abstract
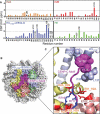
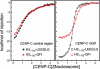
Chromosome segregation during mitosis requires assembly of the kinetochore complex at the centromere. Kinetochore assembly depends on specific recognition of the histone variant CENP-A in the centromeric nucleosome by centromere protein C (CENP-C). We have defined the determinants of this recognition mechanism and discovered that CENP-C binds a hydrophobic region in the CENP-A tail and docks onto the acidic patch of histone H2A and H2B. We further found that the more broadly conserved CENP-C motif uses the same mechanism for CENP-A nucleosome recognition. Our findings reveal a conserved mechanism for protein recruitment to centromeres and a histone recognition mode whereby a disordered peptide binds the histone tail through hydrophobic interactions facilitated by nucleosome docking.
Figures
Comment in
-
An evolving tail of centromere histone variant CENP-A.Cell Cycle. 2013 Oct 1;12(19):3133-4. doi: 10.4161/cc.26353. Epub 2013 Sep 6. Cell Cycle. 2013. PMID: 24013416 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

