BeAtMuSiC: Prediction of changes in protein-protein binding affinity on mutations
- PMID: 23723246
- PMCID: PMC3692068
- DOI: 10.1093/nar/gkt450
BeAtMuSiC: Prediction of changes in protein-protein binding affinity on mutations
Abstract
The ability of proteins to establish highly selective interactions with a variety of (macro)molecular partners is a crucial prerequisite to the realization of their biological functions. The availability of computational tools to evaluate the impact of mutations on protein-protein binding can therefore be valuable in a wide range of industrial and biomedical applications, and help rationalize the consequences of non-synonymous single-nucleotide polymorphisms. BeAtMuSiC (http://babylone.ulb.ac.be/beatmusic) is a coarse-grained predictor of the changes in binding free energy induced by point mutations. It relies on a set of statistical potentials derived from known protein structures, and combines the effect of the mutation on the strength of the interactions at the interface, and on the overall stability of the complex. The BeAtMuSiC server requires as input the structure of the protein-protein complex, and gives the possibility to assess rapidly all possible mutations in a protein chain or at the interface, with predictive performances that are in line with the best current methodologies.
Figures
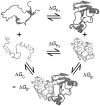
 and
and  are the folding free energies of the two partners of the interaction.
are the folding free energies of the two partners of the interaction.  is the folding free energy of the complex as a whole. In the first binding model, the complex is formed from the association of two individually folded partners, and the binding free energy is
is the folding free energy of the complex as a whole. In the first binding model, the complex is formed from the association of two individually folded partners, and the binding free energy is  . In the second binding model, the proteins are unable to fold independently, and the binding free energy (
. In the second binding model, the proteins are unable to fold independently, and the binding free energy ( ) is thus equal to the folding free energy of the complex (
) is thus equal to the folding free energy of the complex ( ). The figure was made using PyMOL.
). The figure was made using PyMOL.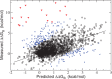
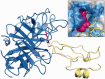
 round of the CAPRI experiment (
round of the CAPRI experiment (
References
-
- Metz A, Ciglia E, Gohlke H. Modulating protein-protein interactions: from structural determinants of binding to druggability prediction to application. Curr. Pharm. Des. 2012;18:4630–4647. - PubMed
-
- Glaser F, Steinberg DM, Vakser IA, Ben-Tal N. Residue frequencies and pairing preferences at protein-protein interfaces. Proteins. 2001;43:89–102. - PubMed
-
- Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

