Effect of CYP3A5 expression on the inhibition of CYP3A-catalyzed drug metabolism: impact on modeling CYP3A-mediated drug-drug interactions
- PMID: 23723360
- PMCID: PMC3716306
- DOI: 10.1124/dmd.112.049940
Effect of CYP3A5 expression on the inhibition of CYP3A-catalyzed drug metabolism: impact on modeling CYP3A-mediated drug-drug interactions
Abstract
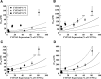
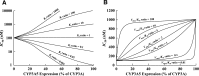
The purpose of this study was to determine the impact of CYP3A5 expression on inhibitory potency (Ki or IC50 values) of CYP3A inhibitors, using recombinant CYP3A4 and CYP3A5 (rCYP3A4 and rCYP3A5) and CYP3A5 genotyped human liver microsomes (HLMs). IC50 ratios between rCYP3A4 and rCYP3A5 (rCYP3A5/rCYP3A4) of ketoconazole (KTZ) and itraconazole (ITZ) were 8.5 and 8.8 for midazolam (MDZ), 4.7 and 9.1 for testosterone (TST), 1.3 and 2.8 for terfenadine, and 0.6 and 1.7 for vincristine, respectively, suggesting substrate- and inhibitor-dependent selectivity of the two azoles. Due to the difference in the IC50 values for CYP3A4 and CYP3A5, nonconcordant expression of CYP3A4 and CYP3A5 protein can significantly affect the observed magnitude of CYP3A-mediated drug-drug interactions in humans. Indeed, the IC50 values of KTZ and ITZ for CYP3A-catalyzed MDZ and TST metabolism were significantly higher in HLMs with CYP3A5*1/*1 and CYP3A5*1/*3 genotypes than in HLMs with the CYP3A5*3/*3 genotype, showing CYP3A5 expression-dependent IC50 values. Moreover, when IC50 values of KTZ and ITZ for different HLMs were kinetically simulated based on CYP3A5 expression level and enzyme-specific IC50 values, a good correlation between the simulated and the experimentally measured IC50 values was observed. Further simulation analysis revealed that both the Ki ratio (for inhibitors) and Vmax/Km ratio (for substrates) between CYP3A4 and CYP3A5 were major factors for CYP3A5 expression-dependent IC50 values. In conclusion, the present study demonstrates that CYP3A5 genotype and expression level have a significant impact on inhibitory potency for CYP3A-catalyzed drug metabolism, but that the magnitude of its effect is inhibitor-substrate pair specific.
Figures
References
-
- Barry A, Levine M. (2010) A systematic review of the effect of CYP3A5 genotype on the apparent oral clearance of tacrolimus in renal transplant recipients. Ther Drug Monit 32:708–714 - PubMed
-
- Chandel N, Aggarwal PK, Minz M, Sakhuja V, Kohli KK, Jha V. (2009) CYP3A5*1/*3 genotype influences the blood concentration of tacrolimus in response to metabolic inhibition by ketoconazole. Pharmacogenet Genomics 19:458–463 - PubMed
-
- Choi MH, Skipper PL, Wishnok JS, Tannenbaum SR. (2005) Characterization of testosterone 11 beta-hydroxylation catalyzed by human liver microsomal cytochromes P450. Drug Metab Dispos 33:714–718 - PubMed
-
- Christensen H, Hestad AL, Molden E, Mathiesen L. (2011) CYP3A5-mediated metabolism of midazolam in recombinant systems is highly sensitive to NADPH-cytochrome P450 reductase activity. Xenobiotica 41:1–5 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

