An overview of autophagy: morphology, mechanism, and regulation
- PMID: 23725295
- PMCID: PMC3894687
- DOI: 10.1089/ars.2013.5371
An overview of autophagy: morphology, mechanism, and regulation
Abstract
Significance: Autophagy is a highly conserved eukaryotic cellular recycling process. Through the degradation of cytoplasmic organelles, proteins, and macromolecules, and the recycling of the breakdown products, autophagy plays important roles in cell survival and maintenance. Accordingly, dysfunction of this process contributes to the pathologies of many human diseases.
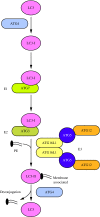
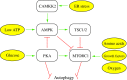
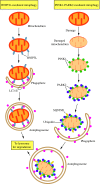
Recent advances: Extensive research is currently being done to better understand the process of autophagy. In this review, we describe current knowledge of the morphology, molecular mechanism, and regulation of mammalian autophagy.
Critical issues: At the mechanistic and regulatory levels, there are still many unanswered questions and points of confusion that have yet to be resolved.
Future directions: Through further research, a more complete and accurate picture of the molecular mechanism and regulation of autophagy will not only strengthen our understanding of this significant cellular process, but will aid in the development of new treatments for human diseases in which autophagy is not functioning properly.
Figures

References
-
- Agarraberes FA. and Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci 114: 2491–2499, 2001 - PubMed
-
- Arsham AM, Howell JJ, and Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem 278: 29655–29660, 2003 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

