Viral pathogen discovery
- PMID: 23725672
- PMCID: PMC5964995
- DOI: 10.1016/j.mib.2013.05.001
Viral pathogen discovery
Abstract
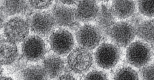
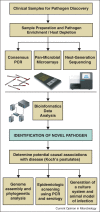
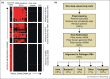
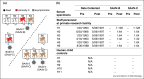
Viral pathogen discovery is of critical importance to clinical microbiology, infectious diseases, and public health. Genomic approaches for pathogen discovery, including consensus polymerase chain reaction (PCR), microarrays, and unbiased next-generation sequencing (NGS), have the capacity to comprehensively identify novel microbes present in clinical samples. Although numerous challenges remain to be addressed, including the bioinformatics analysis and interpretation of large datasets, these technologies have been successful in rapidly identifying emerging outbreak threats, screening vaccines and other biological products for microbial contamination, and discovering novel viruses associated with both acute and chronic illnesses. Downstream studies such as genome assembly, epidemiologic screening, and a culture system or animal model of infection are necessary to establish an association of a candidate pathogen with disease.
Copyright © 2013 The Author. Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Nichol S.T., Spiropoulou C.F., Morzunov S., Rollin P.E., Ksiazek T.G., Feldmann H., Sanchez A., Childs J., Zaki S., Peters C.J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. - PubMed
-
- Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J., Gubareva L.V., Xu X., Bridges C.B., Uyeki T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. - PubMed
-
- Shinde V., Bridges C.B., Uyeki T.M., Shu B., Balish A., Xu X., Lindstrom S., Gubareva L.V., Deyde V., Garten R.J. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. - PubMed
-
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

