Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons
- PMID: 23727006
- PMCID: PMC3884001
- DOI: 10.1016/j.neuroscience.2013.05.042
Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons
Abstract
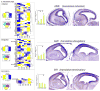
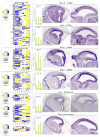
The mature neocortex is a unique six-layered mammalian brain region. It is composed of morphologically and functionally distinct subpopulations of primary projection neurons that form complex circuits across the central nervous system. The precisely-timed generation of projection neurons from neural stem cells governs their differentiation, postmitotic specification, and signaling, and is critical for cognitive and sensorimotor ability. Developmental perturbations to the birthdate, location, and connectivity of neocortical neurons are observed in neurological and psychiatric disorders. These facts are highlighting the importance of the precise spatiotemporal development of the neocortex regulated by intricate transcriptional, but also complex post-transcriptional events. Indeed, mRNA transcripts undergo many post-transcriptional regulatory steps before the production of functional proteins, which specify neocortical neural stem cells and subpopulations of neocortical neurons. Therefore, particular attention is paid to the differential post-transcriptional regulation of key transcripts by RNA-binding proteins, including splicing, localization, stability, and translation. We also present a transcriptome screen of candidate molecules associated with post-transcriptional mRNA processing that are differentially expressed at key developmental time points across neocortical prenatal neurogenesis.
Keywords: RNA-binding protein; neocortex; neural stem cell; post-transcriptional processing; projection pyramidal neuron; translation.
Copyright © 2013 IBRO. Published by Elsevier Ltd. All rights reserved.
Figures




References
-
- Brain Museum. http://brainmuseum.org/
-
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. - PubMed
-
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JDW. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

