Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo
- PMID: 23727241
- PMCID: PMC3700580
- DOI: 10.1016/j.celrep.2013.04.032
Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo
Abstract
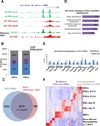
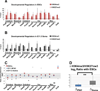
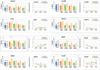
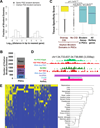
Developmental regulatory genes have both activating (H3K4me3) and repressive (H3K27me3) histone modifications in embryonic stem cells (ESCs). This bivalent configuration is thought to maintain lineage commitment programs in a poised state. However, establishing physiological relevance has been complicated by the high number of cells required for chromatin immunoprecipitation (ChIP). We developed a low-cell-number chromatin immunoprecipitation (low-cell ChIP) protocol to investigate the chromatin of mouse primordial germ cells (PGCs). Genome-wide analysis of embryonic day 11.5 (E11.5) PGCs revealed H3K4me3/H3K27me3 bivalent domains highly enriched at developmental regulatory genes in a manner remarkably similar to ESCs. Developmental regulators remain bivalent and transcriptionally silent through the initiation of sexual differentiation at E13.5. We also identified >2,500 "orphan" bivalent domains that are distal to known genes and expressed in a tissue-specific manner but silent in PGCs. Our results demonstrate the existence of bivalent domains in the germline and raise the possibility that the somatic program is continuously maintained as bivalent, potentially imparting transgenerational epigenetic inheritance.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nature Cell Biology. 2006;8:532–538. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

