The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals
- PMID: 23728292
- PMCID: PMC3702670
- DOI: 10.1038/nsmb.2595
The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals
Abstract
Glucocorticoid receptor (GR) binds to genomic response elements and regulates gene transcription with cell and gene specificity. Within a response element, the precise sequence to which the receptor binds has been implicated in directing its structure and activity. Here, we use NMR chemical-shift difference mapping to show that nonspecific interactions with bases at particular positions in the binding sequence, such as those of the 'spacer', affect the conformation of distinct regions of the rat GR DNA-binding domain. These regions include the DNA-binding surface, the 'lever arm' and the dimerization interface, suggesting an allosteric pathway that signals between the DNA-binding sequence and the associated dimer partner. Disrupting this pathway by mutating the dimer interface alters sequence-specific conformations, DNA-binding kinetics and transcriptional activity. Our study demonstrates that GR dimer partners collaborate to read DNA shape and to direct sequence-specific gene activity.
Figures



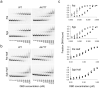


References
-
- Yamamoto K, Darimont B, Wagner R, Iniguez-Lluhi J. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symposia on Quantitative Biology. 1998;63:587–598. - PubMed
-
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Development. 2006;20:1405–1428. - PubMed
-
- Bledsoe RK, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 CA149088/CA/NCI NIH HHS/United States
- R01 CA020535/CA/NCI NIH HHS/United States
- T32 GM008537/GM/NIGMS NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- T32 GM008284/GM/NIGMS NIH HHS/United States
- T32 DK007762/DK/NIDDK NIH HHS/United States
- CA020535/CA/NCI NIH HHS/United States
- 5K99CA149088/CA/NCI NIH HHS/United States
- K99 CA149088/CA/NCI NIH HHS/United States
- K24 DK060617/DK/NIDDK NIH HHS/United States
- GM08537/GM/NIGMS NIH HHS/United States
- T32GM008284/GM/NIGMS NIH HHS/United States
- R37 CA020535/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

