Characterization of Dak Nong virus, an insect nidovirus isolated from Culex mosquitoes in Vietnam
- PMID: 23728735
- PMCID: PMC7087109
- DOI: 10.1007/s00705-013-1741-4
Characterization of Dak Nong virus, an insect nidovirus isolated from Culex mosquitoes in Vietnam
Abstract
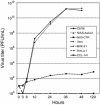
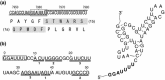
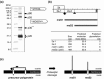
In this study, we isolated and characterized an insect nidovirus from the mosquito Culex tritaeniorhynchus Giles (Diptera: Culicidae) in Vietnam, as an additional member of the new family Mesoniviridae in the order Nidovirales. The virus, designated "Dak Nong virus (DKNV)," shared many characteristics with Cavally virus and Nam Dinh virus, which have also been discovered recently in mosquitoes, and these viruses should be considered members of a single virus species, Alphamesonivirus 1. DKNV grew in cultured mosquito cells but could not replicate in the cultured vertebrate cells tested. N-terminal sequencing of the DKNV structural proteins revealed two posttranslational cleavage sites in the spike glycoprotein precursor. DKNV is assumed to be a new member of the species Alphamesonivirus 1, and the current study provides further understanding of viruses belonging to the new family Mesoniviridae.
Figures




References
-
- de Groot RJ, Cowley JA, Enjuanes L, Faaberg KS, Perlman S, Rottier PJM, Snijder EJ, Ziebuhr J, Gorbalenya AE. Order Nidovirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy; Ninth Report of the International Committee on Taxonomy of Viruses. London: Elsevier; 2012. pp. 785–795.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

