Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes
- PMID: 23729632
- PMCID: PMC3730342
- DOI: 10.1093/gbe/evt086
Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes
Abstract
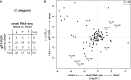
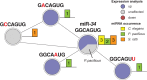
Animal development is complex yet surprisingly robust. Animals may develop alternative phenotypes conditional on environmental changes. Under unfavorable conditions, Caenorhabditis elegans larvae enter the dauer stage, a developmentally arrested, long-lived, and stress-resistant state. Dauer larvae of free-living nematodes and infective larvae of parasitic nematodes share many traits including a conserved endocrine signaling module (DA/DAF-12), which is essential for the formation of dauer and infective larvae. We speculated that conserved post-transcriptional regulatory mechanism might also be involved in executing the dauer and infective larvae fate. We used an unbiased sequencing strategy to characterize the microRNA (miRNA) gene complement in C. elegans, Pristionchus pacificus, and Strongyloides ratti. Our study raised the number of described miRNA genes to 257 for C. elegans, tripled the known gene set for P. pacificus to 362 miRNAs, and is the first to describe miRNAs in a Strongyloides parasite. Moreover, we found a limited core set of 24 conserved miRNA families in all three species. Interestingly, our estimated expression fold changes between dauer versus nondauer stages and infective larvae versus free-living stages reveal that despite the speed of miRNA gene set evolution in nematodes, homologous gene families with conserved "dauer-infective" expression signatures are present. These findings suggest that common post-transcriptional regulatory mechanisms are at work and that the same miRNA families play important roles in developmental arrest and long-term survival in free-living and parasitic nematodes.
Keywords: dauer larvae; microRNA; nematodes; parasites; post-transcriptional regulation.
Figures



References
-
- Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

