Implication of the VirD4 coupling protein of the Lvh type 4 secretion system in virulence phenotypes of Legionella pneumophila
- PMID: 23729650
- PMCID: PMC3719543
- DOI: 10.1128/JB.00430-13
Implication of the VirD4 coupling protein of the Lvh type 4 secretion system in virulence phenotypes of Legionella pneumophila
Abstract
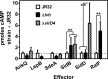
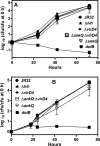
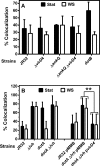
The genome of the Philadelphia-1 strain of Legionella pneumophila, the causative organism of Legionnaires' disease, encodes two virulence-associated type 4 secretion systems (T4SSs), the Dot/Icm type 4B (T4BSS) and the Lvh type 4A (T4ASS). Broth stationary-phase cultures of most dot/icm mutants are defective in entry and evasion of phagosome acidification. However, those virulence defects can be reversed by incubating broth cultures of dot/icm mutants in water, termed water stress (WS). WS reversal requires the lvh T4ASS locus, suggesting an interaction between the two T4SSs in producing Legionella virulence phenotypes. In the current work, the loss of WS reversal in a dotA Δlvh mutant of strain JR32 was shown to be attributable to loss of the lvh virD4 gene, encoding the putative coupling protein of the T4ASS. Transformation of a dotA Δlvh mutant with virD4 also reversed entry and phagosome acidification defects in broth cultures. In addition, broth cultures of Δlvh and ΔvirD4 mutants, which were dot/icm(+), showed 5-fold and >6-fold increases in translocation of the Dot/Icm translocation substrates, proteins RalF and SidD, respectively. These data demonstrate that the Lvh T4ASS functions in both broth stationary-phase cultures conventionally used for infection and cultures exposed to WS treatment. Our studies in a dotA Δlvh mutant and in a dot/icm(+) background establish that VirD4 and the Lvh T4ASS contribute to virulence phenotypes and are consistent with independent functioning of Dot/Icm and Lvh T4SSs or functional substitution of the Lvh VirD4 protein for a component(s) of the Dot/Icm T4BSS.
Figures
Similar articles
-
Implication of proteins containing tetratricopeptide repeats in conditional virulence phenotypes of Legionella pneumophila.J Bacteriol. 2012 Jul;194(14):3579-88. doi: 10.1128/JB.00399-12. Epub 2012 May 4. J Bacteriol. 2012. PMID: 22563053 Free PMC article.
-
Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila.Infect Immun. 2007 Feb;75(2):723-35. doi: 10.1128/IAI.00956-06. Epub 2006 Nov 13. Infect Immun. 2007. PMID: 17101653 Free PMC article.
-
Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins.J Bacteriol. 2010 Nov;192(22):6001-16. doi: 10.1128/JB.00778-10. Epub 2010 Sep 10. J Bacteriol. 2010. PMID: 20833813 Free PMC article.
-
Legionella pneumophila Dot/Icm translocated substrates: a sum of parts.Curr Opin Microbiol. 2009 Feb;12(1):67-73. doi: 10.1016/j.mib.2008.12.004. Epub 2009 Jan 20. Curr Opin Microbiol. 2009. PMID: 19157961 Free PMC article. Review.
-
Effector translocation by the Legionella Dot/Icm type IV secretion system.Curr Top Microbiol Immunol. 2013;376:103-15. doi: 10.1007/82_2013_345. Curr Top Microbiol Immunol. 2013. PMID: 23918176 Review.
Cited by
-
DeepSecE: A Deep-Learning-Based Framework for Multiclass Prediction of Secreted Proteins in Gram-Negative Bacteria.Research (Wash D C). 2023 Oct 25;6:0258. doi: 10.34133/research.0258. eCollection 2023. Research (Wash D C). 2023. PMID: 37886621 Free PMC article.
-
Legionnaires' Disease Mortality in Guinea Pigs Involves the p45 Mobile Genomic Element.J Infect Dis. 2019 Oct 8;220(10):1700-1710. doi: 10.1093/infdis/jiz340. J Infect Dis. 2019. PMID: 31268152 Free PMC article.
-
Genome Sequencing Links Persistent Outbreak of Legionellosis in Sydney (New South Wales, Australia) to an Emerging Clone of Legionella pneumophila Sequence Type 211.Appl Environ Microbiol. 2018 Feb 14;84(5):e02020-17. doi: 10.1128/AEM.02020-17. Print 2018 Mar 1. Appl Environ Microbiol. 2018. PMID: 29247056 Free PMC article.
-
Concept about the Virulence Factor of Legionella.Microorganisms. 2022 Dec 27;11(1):74. doi: 10.3390/microorganisms11010074. Microorganisms. 2022. PMID: 36677366 Free PMC article. Review.
-
Functional type 1 secretion system involved in Legionella pneumophila virulence.J Bacteriol. 2015 Feb;197(3):563-71. doi: 10.1128/JB.02164-14. Epub 2014 Nov 24. J Bacteriol. 2015. PMID: 25422301 Free PMC article.
References
-
- Centers for Disease Control and Prevention 2011. Legionellosis—United States, 2000-2009. MMWR Morb. Mortal. Wkly. Rep. 60:1083–1086 - PubMed
-
- Steinert M, Hentschel U, Hacker J. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149–162 - PubMed
-
- Ge J, Shao F. 2011. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell. Microbiol. 13:1870–1880 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

