Epigenetic layers and players underlying neurodevelopment
- PMID: 23731492
- PMCID: PMC3735843
- DOI: 10.1016/j.tins.2013.05.001
Epigenetic layers and players underlying neurodevelopment
Abstract
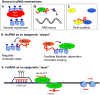
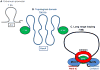
Epigenetic mechanisms convey information above and beyond the sequence of DNA, so it is predicted that they are critical in the complex regulation of brain development and explain the long-lived effects of environmental cues on pre- and early post-natal brain development. Neurons have a complex epigenetic landscape that changes dynamically with transcriptional activity in early life. Here, we summarize progress in our understanding of the discrete layers of the dynamic methylome, chromatin proteome, noncoding RNAs, chromatin loops, and long-range interactions in neuronal development and maturation. Many neurodevelopmental disorders have genetic alterations in these epigenetic modifications or regulators, and these human genetics lessons have demonstrated the importance of these epigenetic players and the epigenetic layers that transcriptional events lay down in the early brain.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Rakyan VK, et al. Metastable epialleles in mammals. Trends in genetics: TIG. 2002;18:348–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

