The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color
- PMID: 23731509
- PMCID: PMC4053823
- DOI: 10.1186/gb-2013-14-6-r53
The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color
Abstract
Background: Theobroma cacao L. cultivar Matina 1-6 belongs to the most cultivated cacao type. The availability of its genome sequence and methods for identifying genes responsible for important cacao traits will aid cacao researchers and breeders.
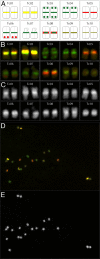
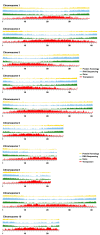
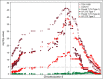
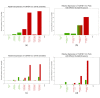
Results: We describe the sequencing and assembly of the genome of Theobroma cacao L. cultivar Matina 1-6. The genome of the Matina 1-6 cultivar is 445 Mbp, which is significantly larger than a sequenced Criollo cultivar, and more typical of other cultivars. The chromosome-scale assembly, version 1.1, contains 711 scaffolds covering 346.0 Mbp, with a contig N50 of 84.4 kbp, a scaffold N50 of 34.4 Mbp, and an evidence-based gene set of 29,408 loci. Version 1.1 has 10x the scaffold N50 and 4x the contig N50 as Criollo, and includes 111 Mb more anchored sequence. The version 1.1 assembly has 4.4% gap sequence, while Criollo has 10.9%. Through a combination of haplotype, association mapping and gene expression analyses, we leverage this robust reference genome to identify a promising candidate gene responsible for pod color variation. We demonstrate that green/red pod color in cacao is likely regulated by the R2R3 MYB transcription factor TcMYB113, homologs of which determine pigmentation in Rosaceae, Solanaceae, and Brassicaceae. One SNP within the target site for a highly conserved trans-acting siRNA in dicots, found within TcMYB113, seems to affect transcript levels of this gene and therefore pod color variation.
Conclusions: We report a high-quality sequence and annotation of Theobroma cacao L. and demonstrate its utility in identifying candidate genes regulating traits.
Figures


References
-
- Figueira A AL. In: Biotechnology of fruit and nut crops. Litz RE, editor. CAB International Biosciences: Wallingford, UK; 2005. Theobroma cacao (Cacao). pp. 639–670.
-
- Foundation TWC. The World Cocoa Foundation. http://www.worldcocoafoundation.org/learn-about-cocoa/
-
- Guiltinan MJ VJ, Zhang D, Figueira A. In: Genomics of Tropical Crop Plants. Moore PH & Ming R, editor. Springer New York; 2008. Genomics of Theobroma cacao, "The Food of the Gods". pp. 146–170.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

