Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease
- PMID: 23731541
- PMCID: PMC3675261
- DOI: 10.1016/j.ajhg.2013.05.002
Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease
Abstract
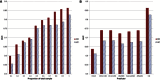
We performed risk assessment for Crohn's disease (CD) and ulcerative colitis (UC), the two common forms of inflammatory bowel disease (IBD), by using data from the International IBD Genetics Consortium's Immunochip project. This data set contains ~17,000 CD cases, ~13,000 UC cases, and ~22,000 controls from 15 European countries typed on the Immunochip. This custom chip provides a more comprehensive catalog of the most promising candidate variants by picking up the remaining common variants and certain rare variants that were missed in the first generation of GWAS. Given this unprecedented large sample size and wide variant spectrum, we employed the most recent machine-learning techniques to build optimal predictive models. Our final predictive models achieved areas under the curve (AUCs) of 0.86 and 0.83 for CD and UC, respectively, in an independent evaluation. To our knowledge, this is the best prediction performance ever reported for CD and UC to date.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. - PMC - PubMed
-
- Evans D.M., Visscher P.M., Wray N.R. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum. Mol. Genet. 2009;18:3525–3531. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

