Genome dynamics in Legionella: the basis of versatility and adaptation to intracellular replication
- PMID: 23732852
- PMCID: PMC3662349
- DOI: 10.1101/cshperspect.a009993
Genome dynamics in Legionella: the basis of versatility and adaptation to intracellular replication
Abstract
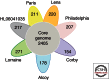
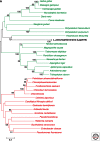
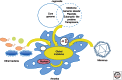
Legionella pneumophila is a bacterial pathogen present in aquatic environments that can cause a severe pneumonia called Legionnaires' disease. Soon after its recognition, it was shown that Legionella replicates inside amoeba, suggesting that bacteria replicating in environmental protozoa are able to exploit conserved signaling pathways in human phagocytic cells. Comparative, evolutionary, and functional genomics suggests that the Legionella-amoeba interaction has shaped this pathogen more than previously thought. A complex evolutionary scenario involving mobile genetic elements, type IV secretion systems, and horizontal gene transfer among Legionella, amoeba, and other organisms seems to take place. This long-lasting coevolution led to the development of very sophisticated virulence strategies and a high level of temporal and spatial fine-tuning of bacteria host-cell interactions. We will discuss current knowledge of the evolution of virulence of Legionella from a genomics perspective and propose our vision of the emergence of this human pathogen from the environment.
Figures

References
-
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140 - PubMed
-
- Atlan D, Coupat-Goutaland B, Risler A, Reyrolle M, Souchon M, Briolay J, Jarraud S, Doublet P, Pélandakis M 2012. Micriamoeba tesseris nov. gen. nov. sp: A new taxon of free-living small-sized Amoebae non-permissive to virulent Legionellae. Protist 163: 888–902 - PubMed
-
- Berger KH, Isberg RR 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7: 7–19 - PubMed
-
- Bruggemann H, Cazalet C, Buchrieser C 2006. Adaptation of Legionella pneumophila to the host environment: Role of protein secretion, effectors and eukaryotic-like proteins. Curr Opin Microbiol 9: 86–94 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
