Autoantibody profiling in multiple sclerosis using arrays of human protein fragments
- PMID: 23732997
- PMCID: PMC3769337
- DOI: 10.1074/mcp.M112.026757
Autoantibody profiling in multiple sclerosis using arrays of human protein fragments
Abstract
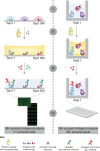
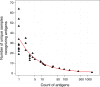
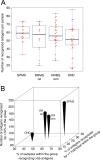
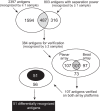
Profiling the autoantibody repertoire with large antigen collections is emerging as a powerful tool for the identification of biomarkers for autoimmune diseases. Here, a systematic and undirected approach was taken to screen for profiles of IgG in human plasma from 90 individuals with multiple sclerosis related diagnoses. Reactivity pattern of 11,520 protein fragments (representing ∼38% of all human protein encoding genes) were generated on planar protein microarrays built within the Human Protein Atlas. For more than 2,000 antigens IgG reactivity was observed, among which 64% were found only in single individuals. We used reactivity distributions among multiple sclerosis subgroups to select 384 antigens, which were then re-evaluated on planar microarrays, corroborated with suspension bead arrays in a larger cohort (n = 376) and confirmed for specificity in inhibition assays. Among the heterogeneous pattern within and across multiple sclerosis subtypes, differences in recognition frequencies were found for 51 antigens, which were enriched for proteins of transcriptional regulation. In conclusion, using protein fragments and complementary high-throughput protein array platforms facilitated an alternative route to discovery and verification of potentially disease-associated autoimmunity signatures, that are now proposed as additional antigens for large-scale validation studies across multiple sclerosis biobanks.
Figures



References
-
- Goodnow C. C., Sprent J., Fazekas, de St Groth B., Vinuesa C. G. (2005) Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435, 590–597 - PubMed
-
- Selmi C. (2011) Autoimmunity in 2010. Autoimmun. Rev. 10, 725–732 - PubMed
-
- Hueber W., Robinson W. H. (2006) Proteomic biomarkers for autoimmune disease. Proteomics 6, 4100–4105 - PubMed
-
- Gibson D. S., Banha J., Penque D., Costa L., Conrads T. P., Cahill D. J., O'Brien J. K., Rooney M. E. (2010) Diagnostic and prognostic biomarker discovery strategies for autoimmune disorders. J. Proteomics 73, 1045–1060 - PubMed
-
- Tjalsma H., Schaeps R. M., Swinkels D. W. (2008) Immunoproteomics: From biomarker discovery to diagnostic applications. Proteomics Clin. Appl. 2, 167–180 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

