A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins
- PMID: 23733182
- PMCID: PMC3711307
- DOI: 10.1074/jbc.M112.446344
A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins
Abstract
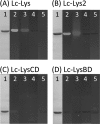
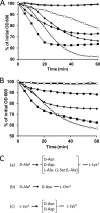
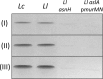
Peptidoglycan hydrolases (PGHs) are responsible for bacterial cell lysis. Most PGHs have a modular structure comprising a catalytic domain and a cell wall-binding domain (CWBD). PGHs of bacteriophage origin, called endolysins, are involved in bacterial lysis at the end of the infection cycle. We have characterized two endolysins, Lc-Lys and Lc-Lys-2, identified in prophages present in the genome of Lactobacillus casei BL23. These two enzymes have different catalytic domains but similar putative C-terminal CWBDs. By analyzing purified peptidoglycan (PG) degradation products, we showed that Lc-Lys is an N-acetylmuramoyl-L-alanine amidase, whereas Lc-Lys-2 is a γ-D-glutamyl-L-lysyl endopeptidase. Remarkably, both lysins were able to lyse only Gram-positive bacterial strains that possess PG with D-Ala(4)→D-Asx-L-Lys(3) in their cross-bridge, such as Lactococcus casei, Lactococcus lactis, and Enterococcus faecium. By testing a panel of L. lactis cell wall mutants, we observed that Lc-Lys and Lc-Lys-2 were not able to lyse mutants with a modified PG cross-bridge, constituting D-Ala(4)→L-Ala-(L-Ala/L-Ser)-L-Lys(3); moreover, they do not lyse the L. lactis mutant containing only the nonamidated D-Asp cross-bridge, i.e. D-Ala(4)→D-Asp-L-Lys(3). In contrast, Lc-Lys could lyse the ampicillin-resistant E. faecium mutant with 3→3 L-Lys(3)-D-Asn-L-Lys(3) bridges replacing the wild-type 4→3 D-Ala(4)-D-Asn-L-Lys(3) bridges. We showed that the C-terminal CWBD of Lc-Lys binds PG containing mainly D-Asn but not PG with only the nonamidated D-Asp-containing cross-bridge, indicating that the CWBD confers to Lc-Lys its narrow specificity. In conclusion, the CWBD characterized in this study is a novel type of PG-binding domain targeting specifically the D-Asn interpeptide bridge of PG.
Keywords: Bacteria; Bacteriophage; Cell Wall; Cell Wall-binding Domain; Endolysin; Hydrolases; Lactobacillus; Lysis; Peptidoglycan; Peptidoglycan Cross-bridge.
Figures


Similar articles
-
Analysis of the peptidoglycan hydrolase complement of Lactobacillus casei and characterization of the major γ-D-glutamyl-L-lysyl-endopeptidase.PLoS One. 2012;7(2):e32301. doi: 10.1371/journal.pone.0032301. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384208 Free PMC article.
-
Identification of an essential gene responsible for D-Asp incorporation in the Lactococcus lactis peptidoglycan crossbridge.Mol Microbiol. 2006 Dec;62(6):1713-24. doi: 10.1111/j.1365-2958.2006.05474.x. Mol Microbiol. 2006. PMID: 17083466
-
Gp29 LysA of mycobacteriophage TM4 can hydrolyze peptidoglycan through an N-acetyl-muramoyl-L-alanine amidase activity.Biochim Biophys Acta Proteins Proteom. 2022 Feb 1;1870(2):140745. doi: 10.1016/j.bbapap.2021.140745. Epub 2021 Dec 11. Biochim Biophys Acta Proteins Proteom. 2022. PMID: 34906734
-
More than just lysins: peptidoglycan hydrolases tailor the cell wall.Curr Opin Microbiol. 2011 Dec;14(6):698-703. doi: 10.1016/j.mib.2011.10.003. Epub 2011 Nov 3. Curr Opin Microbiol. 2011. PMID: 22055466 Free PMC article. Review.
-
Endolysins as antimicrobials.Adv Virus Res. 2012;83:299-365. doi: 10.1016/B978-0-12-394438-2.00007-4. Adv Virus Res. 2012. PMID: 22748813 Review.
Cited by
-
Spontaneous Prophage Induction Contributes to the Production of Membrane Vesicles by the Gram-Positive Bacterium Lacticaseibacillus casei BL23.mBio. 2022 Oct 26;13(5):e0237522. doi: 10.1128/mbio.02375-22. Epub 2022 Oct 6. mBio. 2022. PMID: 36200778 Free PMC article.
-
Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages.Front Microbiol. 2014 May 22;5:236. doi: 10.3389/fmicb.2014.00236. eCollection 2014. Front Microbiol. 2014. PMID: 24904550 Free PMC article. Review.
-
Development of a Magnetic Bead-Based Method for Specific Detection of Enterococcus faecalis Using C-Terminal Domain of ECP3 Phage Endolysin.J Microbiol Biotechnol. 2023 Jul 28;33(7):964-972. doi: 10.4014/jmb.2302.02033. Epub 2023 Apr 20. J Microbiol Biotechnol. 2023. PMID: 37164751 Free PMC article.
-
Regulation of Cell Wall Plasticity by Nucleotide Metabolism in Lactococcus lactis.J Biol Chem. 2016 May 20;291(21):11323-36. doi: 10.1074/jbc.M116.714303. Epub 2016 Mar 28. J Biol Chem. 2016. PMID: 27022026 Free PMC article.
-
Impact of crossbridge structure on peptidoglycan crosslinking: A synthetic stem peptide approach.Methods Enzymol. 2022;665:259-279. doi: 10.1016/bs.mie.2021.11.019. Epub 2022 Feb 2. Methods Enzymol. 2022. PMID: 35379437 Free PMC article.
References
-
- Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 - PubMed
-
- Chapot-Chartier M.-P. (2010) in Prokaryotic Cell Wall Compounds (König H., Claus H., Varna A., eds) pp. 383–406, Springer Verlag Berlin, Heidelberg, Germany
-
- Loessner M. J. (2005) Bacteriophage endolysins–current state of research and applications. Curr. Opin. Microbiol. 8, 480–487 - PubMed
-
- Hermoso J. A., García J. L., García P. (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 10, 461–472 - PubMed
-
- Wang I. N., Smith D. L., Young R. (2000) Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799–825 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

