MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways
- PMID: 23733368
- PMCID: PMC3721471
- DOI: 10.1002/emmm.201202318
MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways
Abstract
Activation of inflammatory pathways in the endothelium contributes to vascular diseases, including sepsis and atherosclerosis. We demonstrate that miR-146a and miR-146b are induced in endothelial cells upon exposure to pro-inflammatory cytokines. Despite the rapid transcriptional induction of the miR-146a/b loci, which is in part mediated by EGR-3, miR-146a/b induction is delayed and sustained compared to the expression of leukocyte adhesion molecules, and in fact coincides with the down-regulation of inflammatory gene expression. We demonstrate that miR-146 negatively regulates inflammation. Over-expression of miR-146a blunts endothelial activation, while knock-down of miR-146a/b in vitro or deletion of miR-146a in mice has the opposite effect. MiR-146 represses the pro-inflammatory NF-κB pathway as well as the MAP kinase pathway and downstream EGR transcription factors. Finally, we demonstrate that HuR, an RNA binding protein that promotes endothelial activation by suppressing expression of endothelial nitric oxide synthase (eNOS), is a novel miR-146 target. Thus, we uncover an important negative feedback regulatory loop that controls pro-inflammatory signalling in endothelial cells that may impact vascular inflammatory diseases.
Keywords: atherosclerosis; endothelium; gene regulation; inflammation; microRNA.
© 2013 The Authors. Published by John Wiley and Sons, Ltd on behalf of EMBO.
Figures
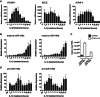
Levels of pro-inflammatory genes (VCAM-1, SELE (E-Selectin), ICAM1) were measured in IL-1β-treated human umbilical vein endothelial cells (HUVEC) by quantitative reverse transcriptase real-time PCR (qRT-PCR), revealing that these inflammatory genes were rapidly induced by IL-1β, but decreased by 24 h (h). Data represent the mean ± SEM of three independent experiments.
Levels of mature miR-146a and miR-146b were assessed by qRT-PCR (n = 3). MiR-146a/b were increased following prolonged treatment with IL-1β.
The copy numbers of miR-146a and miR-146b were quantified in non-stimulated (NS) and 72 h IL-1β-treated endothelial cells (n = 3).
Assessment of the primary transcripts (pri-cursors), pri-miR-146a and pri-miR-146b, by qRT-PCR demonstrated rapid transcriptional up-regulation, which mirrored that of other inflammatory genes (n = 5). The transcription of miR-146a/b appeared to be sustained during prolonged inflammation.
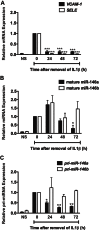

MiR-146a was over-expressed in endothelial cells by transfection of miR-146a mimic and levels of a known target of miR-146, TRAF6, were assessed by Western blot. GAPDH was used as a loading control and densitometry is indicated above. A representative experiment is shown.
Expression of TRAF6 (white bar), inflammatory genes (VCAM-1, ICAM1, SELE (E-Selectin), and MCP-1; black bars), as well as NOS3 (eNOS; grey bar), were measured in unstimulated (left) and IL-1β-stimulated cells (right) by qRT-PCR. For inflammatory genes, gene expression was analysed 1.5 h after addition of IL-1β, while NOS3 was assessed after 8 h. Data is presented as mRNA levels in miR-146a mimic-transfected cells compared to control mimic-transfected cells, with the dotted line indicating a ratio of 1 (i.e., no change; n = 4). p Values (t-test) from left to right are 0.031, 0.023, 0.0002, 0.0001, 0.002, 0.014, 0.006, 0.012, 0.012, 0.006, 0.011 and 0.045, respectively.
Western blotting was performed to measure expression of VCAM-1, E-Selectin and ICAM-1 protein in control and miR-146a mimic-transfected cells. Densitometry is indicated.
Western blotting of eNOS protein was performed in control and miR-146a mimic-transfected cells.
Adhesion of the mononuclear cell line, THP-1, to unstimulated and IL-1β-treated endothelial cells transfected with control or miR-146a mimic was visualized (left) and quantified (right), revealing a strong anti-adhesive effect of miR-146a over-expression. Scale bar is 200 μm. Shown is a representative experiment (mean ± SEM) with three replicate wells and three images per well for each condition. ANOVA, p < 0.0001. ***Indicates a significant difference between IL-1β-treated control and miR-146a mimic-transfected cells, p < 0.001.
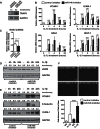
Endothelial cells were transfected with a miR-146 LNA inhibitor (which reduces levels of miR-146a and miR-146b by >80%), and the level of a known target of miR-146, TRAF6, was measured by Western blot.
The expression of inflammatory genes (VCAM-1, ICAM-1, SELE, and MCP-1) in unstimulated and IL-1β-stimulated cells was assessed by qRT-PCR. Data represents mean ± SEM of three independent experiments. Significant p values (t-test) are indicated above.
Levels of NOS3 mRNA were assessed by qRT-PCR in control inhibitor and miR-146 inhibitor transfected cells after 24 h of IL-1β treatment (n = 3). Data is expressed relative to untreated cells.
Levels of eNOS protein were measured in control and miR-146 inhibitor transfected cells.
Western blotting was performed to measure VCAM-1, E-Selectin and ICAM-1 protein expression in control inhibitor and miR-146 inhibitor transfected cells.
Monocyte adhesion assays were performed in control and miR-146 inhibitor transfected endothelial cells. Representative images are shown (above) and quantification of a representative experiment (three replicate wells, three images per well) is shown (below). Scale bar is 200 μm. ANOVA, p < 0.0001. ***Indicates a significant difference between IL-1β-treated control and miR-146 inhibitor-transfected cells, p < 0.001.

The activity of a NF-κB promoter-luciferase reporter construct was assessed in endothelial cells transfected with control mimic, miR-146a mimic, control inhibitor or miR-146 inhibitor. MiR-146a over-expression reduced IL-1β-induced NF-κB-dependent promoter activity, while inhibition of miR-146 enhanced activity. Data represents the mean ± SEM of three independent experiments. ANOVA, p < 0.0001 for mimic and inhibitor data. ** and *** indicate a significant difference between the indicated groups, p < 0.01 and p < 0.001, respectively.
Activation of the MAP kinase pathway was assessed by measuring the levels of phosphorylated ERK (pERK) (p42/p44). Total levels of ERK2 were used as a loading control. MiR-146a over-expression inhibited the basal and IL-1β-induced levels of pERK, while miR-146 inhibitor had the opposite effect.
Induction of EGR-1 and EGR-3 in response to IL-1β was assessed by qRT-PCR, demonstrating rapid and transient induction (n = 3).
MiR-146a over-expression inhibited the IL-1β-mediated induction of EGR-1 and EGR-3, while inhibition of miR-146 enhanced the induction of EGR-3 (n = 3). Significant p values (t-test) from left to right are 0.002, 0.004 and 0.022, respectively.
Schematic of a potential miR-146 binding site in the 3′ UTR of EGR-3 (top). Luciferase assays utilizing wild-type or seed-mutated EGR-3 concatemer or TRAF6 3′ UTR sequences were performed in the presence of control or miR-146a mimic (p = 0.042, t-test, n = 3).
Activation of the JNK/AP-1 pathway was assessed by measuring the induction of c-Fos and c-Jun by qRT-PCR. MiR-146a over-expression reduced c-Fos expression, while inhibition of miR-146 enhanced c-Jun expression in response to IL-1β (n = 4). Significant p values (t-test) from left to right are 0.005, 0.024, respectively.

Treatment of endothelial cells with the MEK inhibitor, U0126, inhibited the basal expression (t-test, p = 0.0003) and IL-1β-dependent induction (t-test, p = 0.037) of EGR-3 (n = 3).
Induction of pri-miR-146a and pri-miR-146b by IL-1β was reduced in cells pre-treated with the MAP kinase inhibitor, U0126. Data represents the relative induction of pri-miR-146a/b in cells treated with U0126 compared to cells treated with DMSO (vehicle) (n = 4). p = 0.037 for pri-miR-146a and p = 0.010 for pri-miR-146b (t-test).
EGR-3 knock-down by siRNA transfection reduced the basal (t-test, p < 0.0001) and IL-1β-induced levels (t-test, p = 0.004) of EGR-3 (n = 5).
The induction of pri-miR-146a and pri-miR-146b was also reduced in EGR-3 knock-down cells (n = 5). p = 0.023 for pri-miR-146a and p = 0.013 for pri-miR-146b (t-test).
Schematic indicating a potential EGR binding site (shaded area) in the miR-146b promoter. Sequence comparison between various species is indicated. Asterisks indicate conserved nucleotides across all species.
Schematic of deletion of the EGR binding site in the miR-146b promoter.
A miR-146b promoter-luciferase reporter was responsive to EGR-3 over-expression (OE) in bovine aortic endothelial cells (BAEC) and mutation of a conserved EGR binding site abrogated this responsiveness. Data depicts the fold induction with EGR-3 OE compared to control. Shown is a representative experiment (n = 3 replicates). p = 0.0017 (t-test).
A miR-146b promoter-luciferase reporter was modestly induced in response to IL-1β and this induction was not observed when the EGR site was mutated. IL-1β was added at concentrations of 10, 20 or 40 ng/mL. Shown is a representative experiment (n = 3 replicates). ANOVA, p = 0.011. * and ** indicate a significant difference between the indicated groups, p < 0.05, p < 0.01, respectively.

Schematic of a potential miR-146 binding site in the 3′ UTR of HuR.
Luciferase assays utilizing wild-type (WT) or seed-mutated (Mut) HuR 3′ UTR sequences were performed in the presence of control or miR-146a mimic (mean ± SEM, p = 0.008, t-test, n = 4).
HuR protein levels were quantified by Western blot in cells transfected with control or miR-146a mimic (left) or control or miR-146 inhibitor (right).
The adhesion of THP-1 cells to vehicle or IL-1β treated cells transfected with control or HuR siRNAs revealed that HuR promotes endothelial activation. A representative experiment is shown (three replicate wells, three images per well). ANOVA, p < 0.0001. ***Indicates a significant decrease in THP-1 adhesion in IL-1β-treated HuR knock-down cells, p < 0.001.
THP-1 adhesion assays were performed with endothelial cells transfected with control or miR-146 inhibitor and control or HuR siRNA. HuR knock-down reduced the elevated adhesion of THP-1 to endothelial cells transfected with miR-146 inhibitor. A representative experiment is shown (three replicate wells, three images per well). ANOVA = 0.016. *Indicates a significant difference between indicated groups, p < 0.05.
Knock-down of HuR (above) or TRAF6 (below) was performed and the induction of adhesion molecules (typified by VCAM-1) and eNOS (NOS3) was assessed by qRT-PCR. Expression of other inflammatory genes is indicated in Supporting Information Fig S9B. HuR knock-down did not reduce the induction of VCAM-1, in contrast to TRAF6 knock-down, which strongly inhibited VCAM-1 induction. However, HuR knock-down significantly elevated levels of NOS3. Shown is the mean ± SEM of three independent experiments. Significant p values (t-test) are indicated above.
Levels of eNOS protein were elevated in HuR knock-down cells, and eNOS was not down-regulated in HuR knock-down cells treated with IL-1β.
The nitric oxide inhibitor,
l -NAME, negated the reduced THP-1 adhesion observed in HuR knock-down cells. A representative experiment is shown (three replicate wells, three images per well). ANOVA, p < 0.0001. ***Indicates a significant difference between groups, p < 0.001.

Endothelial cells and cells in the vessel wall were isolated from the descending aorta of wild-type mice, and expression of miR-126 (as a control for endothelial cells) and miR-146a/b were measured by qRT-PCR. Expression was normalized to U6. MiR-146a was significantly enriched in the endothelium compared to the vessel wall (n = 4). Significant p values (t-test) are indicated above.
Levels of miR-146a and miR-146b were quantified by qRT-PCR in hearts from wild-type and miR-146a−/− mice (3–4 months of age, n = 3). Expression of miR-146a was >6-fold higher than miR-146b and miR-146b expression was not affected by loss of miR-146a.
Expression of HuR mRNA was elevated in the hearts of miR-146a−/− mice as assessed by qRT-PCR (left, p = 0.031, t-test, n = 3). Western blot revealed elevated levels of HuR and Traf6 (right).
Wild-type and miR-146a−/− mice (3–4 months of age, n = 4) were injected with PBS or 125 ng of IL-1β by tail vein injection and hearts were harvested after 2 or 4 h. Expression of inflammatory genes was assessed by qRT-PCR. While basal levels of these genes were unchanged in unstimulated mice (PBS injection), the induction of Vcam-1, Icam-1, Sele, Mcp-1, Egr-1 and Egr-3 was enhanced at 2 h in IL-1β treated mice, and Sele and Icam-1 were still elevated at 4 h. Significant p values (t-test) are indicated above.
Expression of Vcam-1 protein was elevated after a 2 h IL-1β treatment in miR-146a−/− mice compared to wild-type mice.
Localization of Vcam-1 expression was assessed by immunofluorescence, revealing an enhancement of Vcam-1 expression in the endothelium and in puncta adjacent to the endothelium of miR-146a−/− mice treated with IL-1β for 4 h. Scale bars = 20 μm.
Similar articles
-
MicroRNA-146a provides feedback regulation of lyme arthritis but not carditis during infection with Borrelia burgdorferi.PLoS Pathog. 2014 Jun 26;10(6):e1004212. doi: 10.1371/journal.ppat.1004212. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24967703 Free PMC article.
-
Inflammatory gene networks in term human decidual cells define a potential signature for cytokine-mediated parturition.Am J Obstet Gynecol. 2016 Feb;214(2):284.e1-284.e47. doi: 10.1016/j.ajog.2015.08.075. Epub 2015 Sep 5. Am J Obstet Gynecol. 2016. PMID: 26348374
-
Dysregulated expression of miR-146a contributes to age-related dysfunction of macrophages.Aging Cell. 2012 Feb;11(1):29-40. doi: 10.1111/j.1474-9726.2011.00757.x. Epub 2011 Nov 16. Aging Cell. 2012. PMID: 21981419
-
Mechanosensitive microRNAs-role in endothelial responses to shear stress and redox state.Free Radic Biol Med. 2013 Sep;64:61-8. doi: 10.1016/j.freeradbiomed.2013.05.034. Epub 2013 May 30. Free Radic Biol Med. 2013. PMID: 23727269 Free PMC article. Review.
-
Endothelial microRNAs and atherosclerosis.Curr Atheroscler Rep. 2013 Dec;15(12):372. doi: 10.1007/s11883-013-0372-2. Curr Atheroscler Rep. 2013. PMID: 24158362 Free PMC article. Review.
Cited by
-
MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis.Nat Rev Cardiol. 2015 Jun;12(6):361-74. doi: 10.1038/nrcardio.2015.38. Epub 2015 Apr 7. Nat Rev Cardiol. 2015. PMID: 25855604 Review.
-
Anti-atherosclerosis mechanisms associated with regulation of non-coding RNAs by active monomers of traditional Chinese medicine.Front Pharmacol. 2023 Nov 6;14:1283494. doi: 10.3389/fphar.2023.1283494. eCollection 2023. Front Pharmacol. 2023. PMID: 38026969 Free PMC article. Review.
-
MicroRNA Modulation during Orthodontic Tooth Movement: A Promising Strategy for Novel Diagnostic and Personalized Therapeutic Interventions.Int J Mol Sci. 2022 Dec 7;23(24):15501. doi: 10.3390/ijms232415501. Int J Mol Sci. 2022. PMID: 36555142 Free PMC article. Review.
-
Association between polymorphisms in microRNAs and ischemic stroke in an Asian population: evidence based on 6,083 cases and 7,248 controls.Clin Interv Aging. 2018 Sep 12;13:1709-1726. doi: 10.2147/CIA.S174000. eCollection 2018. Clin Interv Aging. 2018. PMID: 30254431 Free PMC article.
-
Leishmania survives by exporting miR-146a from infected to resident cells to subjugate inflammation.Life Sci Alliance. 2022 Feb 24;5(6):e202101229. doi: 10.26508/lsa.202101229. Print 2022 Jun. Life Sci Alliance. 2022. PMID: 35210329 Free PMC article.
References
-
- Aird WC. The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood. 2003;101:3765–3777. - PubMed
-
- Albrecht C, Preusch MR, Hofmann G, Morris-Rosenfeld S, Blessing E, Rosenfeld ME, Katus HA, Bea F. Egr-1 deficiency in bone marrow-derived cells reduces atherosclerotic lesion formation in a hyperlipidaemic mouse model. Cardiovasc Res. 2010;86:321–329. - PubMed
-
- Anderson HD, Rahmutula D, Gardner DG. Tumor necrosis factor-alpha inhibits endothelial nitric-oxide synthase gene promoter activity in bovine aortic endothelial cells. J Biol Chem. 2004;279:963–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

