Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure
- PMID: 23733475
- PMCID: PMC3719707
- DOI: 10.1128/AAC.00263-13
Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure
Abstract
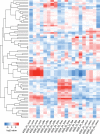
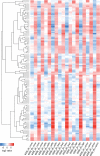
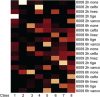
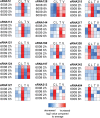
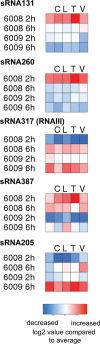
The critical role of noncoding small RNAs (sRNAs) in the bacterial response to changing conditions is increasingly recognized. However, a specific role for sRNAs during antibiotic exposure has not been investigated in Staphylococcus aureus. Here, we used Illumina RNA-Seq to examine the sRNA response of multiresistant sequence type 239 (ST239) S. aureus after exposure to four antibiotics (vancomycin, linezolid, ceftobiprole, and tigecycline) representing the major classes of antimicrobials used to treat methicillin-resistant S. aureus (MRSA) infections. We identified 409 potential sRNAs and then compared global sRNA and mRNA expression profiles at 2 and 6 h, without antibiotic exposure and after exposure to each antibiotic, for a vancomycin-susceptible strain (JKD6009) and a vancomycin-intermediate strain (JKD6008). Exploration of this data set by multivariate analysis using a novel implementation of nonnegative matrix factorization (NMF) revealed very different responses for mRNA and sRNA. Where mRNA responses clustered with strain or growth phase conditions, the sRNA responses were predominantly linked to antibiotic exposure, including sRNA responses that were specific for particular antibiotics. A remarkable feature of the antimicrobial response was the prominence of antisense sRNAs to genes encoding proteins involved in protein synthesis and ribosomal function. This study has defined a large sRNA repertoire in epidemic ST239 MRSA and shown for the first time that a subset of sRNAs are part of a coordinated transcriptional response to specific antimicrobial exposures in S. aureus. These data provide a framework for interrogating the role of staphylococcal sRNAs in antimicrobial resistance and exploring new avenues for sRNA-based antimicrobial therapies.
Figures

References
-
- Turnidge JD, Kotsanas D, Munckhof W, Roberts S, Bennett CM, Nimmo GR, Coombs GW, Murray RJ, Howden B, Johnson PD, Dowling K. 2009. Staphylococcus aureus bacteraemia: a major cause of mortality in Australia and New Zealand. Med. J. Aust. 191:368–373 - PubMed
-
- Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 - PMC - PubMed
-
- Nimmo GR, Pearson JC, Collignon PJ, Christiansen KJ, Coombs GW, Bell JM, McLaws ML. 2007. Prevalence of MRSA among Staphylococcus aureus isolated from hospital inpatients, 2005: report from the Australian Group for Antimicrobial Resistance. Commun. Dis. Intell. 31:288–296 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

