Identity by descent: variation in meiosis, across genomes, and in populations
- PMID: 23733848
- PMCID: PMC3664843
- DOI: 10.1534/genetics.112.148825
Identity by descent: variation in meiosis, across genomes, and in populations
Abstract
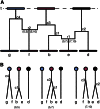
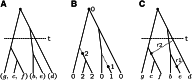
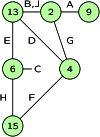
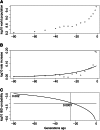
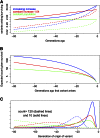
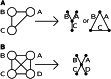
Gene identity by descent (IBD) is a fundamental concept that underlies genetically mediated similarities among relatives. Gene IBD is traced through ancestral meioses and is defined relative to founders of a pedigree, or to some time point or mutational origin in the coalescent of a set of extant genes in a population. The random process underlying changes in the patterns of IBD across the genome is recombination, so the natural context for defining IBD is the ancestral recombination graph (ARG), which specifies the complete ancestry of a collection of chromosomes. The ARG determines both the sequence of coalescent ancestries across the chromosome and the extant segments of DNA descending unbroken by recombination from their most recent common ancestor (MRCA). DNA segments IBD from a recent common ancestor have high probability of being of the same allelic type. Non-IBD DNA is modeled as of independent allelic type, but the population frame of reference for defining allelic independence can vary. Whether of IBD, allelic similarity, or phenotypic covariance, comparisons may be made to other genomic regions of the same gametes, or to the same genomic regions in other sets of gametes or diploid individuals. In this review, I present IBD as the framework connecting evolutionary and coalescent theory with the analysis of genetic data observed on individuals. I focus on the high variance of the processes that determine IBD, its changes across the genome, and its impact on observable data.
Figures
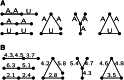
References
-
- Abecasis G. R., Cherny S. S., Cookson W. O., Cardon L. R., 2002. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30: 97–101. - PubMed
-
- Albrechtsen A., Korneliussen T. S., Moltke I., van Overseem Hansen T., Nielsen F. C., et al. , 2009. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet. Epidemiol. 33: 266–274. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

