Prolonged light exposure induces widespread phase shifting in the circadian clock and visual pigment gene expression of the Arvicanthis ansorgei retina
- PMID: 23734075
- PMCID: PMC3668684
Prolonged light exposure induces widespread phase shifting in the circadian clock and visual pigment gene expression of the Arvicanthis ansorgei retina
Abstract
Purpose: Prolonged periods of constant lighting are known to perturb circadian clock function at the molecular, physiological, and behavioral levels. However, the effects of ambient lighting regimes on clock gene expression and clock outputs in retinal photoreceptors--rods, cones and intrinsically photosensitive retinal ganglion cells--are only poorly understood.
Methods: Cone-rich diurnal rodents (Muridae: Arvicanthis ansorgei) were maintained under and entrained to a 12 h:12 h light-dark cycle (LD; light: ~300 lux). Three groups were then examined: control (continued maintenance on LD); animals exposed to a 36 h dark period before sampling over an additional 24 h period of darkness (DD); and animals exposed to a 36 h light period before sampling over an additional 24 h period of light (~300 lux, LL). Animals were killed every 3 or 4 h over 24 h, their retinas dissected, and RNA extracted. Oligonucleotide primers were designed for the Arvicanthis clock genes Per1, Per2, Cry1, Cry2, and Bmal1, and for transcripts specific for rods (rhodopsin), cones (short- and mid-wavelength sensitive cone opsin, cone arrestin, arylalkylamine N-acetyltransferase) and intrinsically photosensitive retinal ganglion cells (melanopsin). Gene expression was analyzed by real-time PCR.
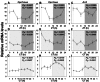
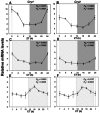
Results: In LD, expression of all genes except cone arrestin was rhythmic and coordinated, with acrophases of most genes at or shortly following the time of lights on (defined as zeitgeber time 0). Arylalkylamine N-acetyltransferase showed maximal expression at zeitgeber time 20. In DD conditions the respective profiles showed similar phase profiles, but were mostly attenuated in amplitude, or in the case of melanopsin, did not retain rhythmic expression. In LL, however, the expression profiles of all clock genes and most putative output genes were greatly altered, with either abolition of daily variation (mid-wavelength cone opsin) or peak expression shifted by 4-10 h.
Conclusions: These data are the first to provide detailed measures of retinal clock gene and putative clock output gene expression in a diurnal mammal, and show the highly disruptive effects of inappropriate (nocturnal) lighting on circadian and photoreceptor gene regulation.
Figures






References
-
- Cermakian N, Sassone-Corsi P. Environmental stimulus perception and control of circadian clocks. Curr Opin Neurobiol. 2002;12:359–65. - PubMed
-
- Tosini G. Melatonin circadian rhythm in the retina of mammals. Chronobiol Int. 2000;17:599–612. - PubMed
-
- Bassi CJ, Powers MK. Circadian rhythm in goldfish visual sensitivity. Invest Ophthalmol Vis Sci. 1987;28:1811–5. - PubMed
-
- Ko GY, Ko ML, Dryer SE. Circadian regulation of cGMP-gated cationic channels of chick retinal cones. Erk MAP Kinase and Ca2+/calmodulin-dependent protein kinase II. Neuron. 2001;29:255–66. - PubMed
-
- von Schantz M, Lucas RJ, Foster RG. Circadian oscillation of photopigment transcript levels in the mouse retina. Brain Res Mol Brain Res. 1999;72:108–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
