Comparative transcriptome profiling reveals different expression patterns in Xanthomonas oryzae pv. oryzae strains with putative virulence-relevant genes
- PMID: 23734193
- PMCID: PMC3667120
- DOI: 10.1371/journal.pone.0064267
Comparative transcriptome profiling reveals different expression patterns in Xanthomonas oryzae pv. oryzae strains with putative virulence-relevant genes
Abstract
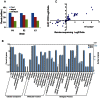
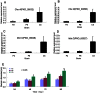
Xanthomonas oryzae pv. oryzae (Xoo) is the causal agent of rice bacterial blight, which is a major rice disease in tropical Asian countries. An attempt has been made to investigate gene expression patterns of three Xoo strains on the minimal medium XOM2, PXO99 (P6) and PXO86 (P2) from the Philippines, and GD1358 (C5) from China, which exhibited different virulence in 30 rice varieties, with putative virulence factors using deep sequencing. In total, 4,781 transcripts were identified in this study, and 1,151 and 3,076 genes were differentially expressed when P6 was compared with P2 and with C5, respectively. Our results indicated that Xoo strains from different regions exhibited distinctly different expression patterns of putative virulence-relevant genes. Interestingly, 40 and 44 genes involved in chemotaxis and motility exhibited higher transcript alterations in C5 compared with P6 and P2, respectively. Most other genes associated with virulence, including exopolysaccharide (EPS) synthesis, Hrp genes and type III effectors, including Xanthomonas outer protein (Xop) effectors and transcription activator-like (TAL) effectors, were down-regulated in C5 compared with P6 and P2. The data were confirmed by real-time quantitative RT-PCR, tests of bacterial motility, and enzyme activity analysis of EPS and xylanase. These results highlight the complexity of Xoo and offer new avenues for improving our understanding of Xoo-rice interactions and the evolution of Xoo virulence.
Conflict of interest statement
Figures


References
-
- NiÑo Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7(5): 303–324. - PubMed
-
- Mew TW (1987) Current status and future prospects of research on bacterial blight of rice. Ann Rev Phytopathol 25: 359–382.
-
- Ochiai H, Inoue Y, Takeya M, Sasaki A, Kaku H (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn Agric Res Q 39(4): 275–287.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

