Functional diversity and structural disorder in the human ubiquitination pathway
- PMID: 23734257
- PMCID: PMC3667038
- DOI: 10.1371/journal.pone.0065443
Functional diversity and structural disorder in the human ubiquitination pathway
Abstract
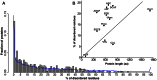
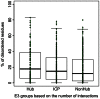
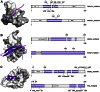
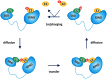
The ubiquitin-proteasome system plays a central role in cellular regulation and protein quality control (PQC). The system is built as a pyramid of increasing complexity, with two E1 (ubiquitin activating), few dozen E2 (ubiquitin conjugating) and several hundred E3 (ubiquitin ligase) enzymes. By collecting and analyzing E3 sequences from the KEGG BRITE database and literature, we assembled a coherent dataset of 563 human E3s and analyzed their various physical features. We found an increase in structural disorder of the system with multiple disorder predictors (IUPred - E1: 5.97%, E2: 17.74%, E3: 20.03%). E3s that can bind E2 and substrate simultaneously (single subunit E3, ssE3) have significantly higher disorder (22.98%) than E3s in which E2 binding (multi RING-finger, mRF, 0.62%), scaffolding (6.01%) and substrate binding (adaptor/substrate recognition subunits, 17.33%) functions are separated. In ssE3s, the disorder was localized in the substrate/adaptor binding domains, whereas the E2-binding RING/HECT-domains were structured. To demonstrate the involvement of disorder in E3 function, we applied normal modes and molecular dynamics analyses to show how a disordered and highly flexible linker in human CBL (an E3 that acts as a regulator of several tyrosine kinase-mediated signalling pathways) facilitates long-range conformational changes bringing substrate and E2-binding domains towards each other and thus assisting in ubiquitin transfer. E3s with multiple interaction partners (as evidenced by data in STRING) also possess elevated levels of disorder (hubs, 22.90% vs. non-hubs, 18.36%). Furthermore, a search in PDB uncovered 21 distinct human E3 interactions, in 7 of which the disordered region of E3s undergoes induced folding (or mutual induced folding) in the presence of the partner. In conclusion, our data highlights the primary role of structural disorder in the functions of E3 ligases that manifests itself in the substrate/adaptor binding functions as well as the mechanism of ubiquitin transfer by long-range conformational transitions.
Conflict of interest statement
Figures



References
-
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991. - PubMed
-
- Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. - PubMed
-
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479. - PubMed
-
- Hochstrasser M (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

