A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo
- PMID: 23735295
- PMCID: PMC3723625
- DOI: 10.1105/tpc.113.111567
A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo
Abstract
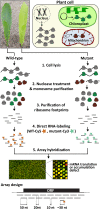
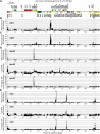
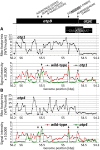
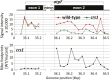
The profiling of ribosome footprints by deep sequencing has revolutionized the analysis of translation by mapping ribosomes with high resolution on a genome-wide scale. We present a variation on this approach that offers a rapid and cost-effective alternative for the genome-wide profiling of chloroplast ribosomes. Ribosome footprints from leaf tissue are hybridized to oligonucleotide tiling microarrays of the plastid ORFeome and report the abundance and translational status of every chloroplast mRNA. Each assay replaces several time-consuming traditional methods while also providing information that was previously inaccessible. To illustrate the utility of the approach, we show that it detects known defects in chloroplast gene expression in several nuclear mutants of maize (Zea mays) and that it reveals previously unsuspected defects. Furthermore, it provided firm answers to several lingering questions in chloroplast gene expression: (1) the overlapping atpB/atpE open reading frames, whose translation had been proposed to be coupled, are translated independently in vivo; (2) splicing is not a prerequisite for translation initiation on an intron-containing chloroplast RNA; and (3) a feedback control mechanism that links the synthesis of ATP synthase subunits in Chlamydomonas reinhardtii does not exist in maize. An analogous approach is likely to be useful for studies of mitochondrial gene expression.
Figures



Similar articles
-
Ribosome Profiling in Maize.Methods Mol Biol. 2018;1676:165-183. doi: 10.1007/978-1-4939-7315-6_10. Methods Mol Biol. 2018. PMID: 28986910
-
Dynamics of Chloroplast Translation during Chloroplast Differentiation in Maize.PLoS Genet. 2016 Jul 14;12(7):e1006106. doi: 10.1371/journal.pgen.1006106. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27414025 Free PMC article.
-
Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane.Proc Natl Acad Sci U S A. 2015 Mar 31;112(13):E1678-87. doi: 10.1073/pnas.1424655112. Epub 2015 Mar 16. Proc Natl Acad Sci U S A. 2015. PMID: 25775549 Free PMC article.
-
Chloroplast Translation: Structural and Functional Organization, Operational Control, and Regulation.Plant Cell. 2018 Apr;30(4):745-770. doi: 10.1105/tpc.18.00016. Epub 2018 Apr 2. Plant Cell. 2018. PMID: 29610211 Free PMC article. Review.
-
Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale.Wiley Interdiscip Rev RNA. 2013 Sep-Oct;4(5):473-90. doi: 10.1002/wrna.1172. Epub 2013 May 20. Wiley Interdiscip Rev RNA. 2013. PMID: 23696005 Free PMC article. Review.
Cited by
-
Seamless editing of the chloroplast genome in plants.BMC Plant Biol. 2016 Jul 29;16(1):168. doi: 10.1186/s12870-016-0857-6. BMC Plant Biol. 2016. PMID: 27474038 Free PMC article.
-
N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men.Mol Cell Proteomics. 2014 May;13(5):1245-61. doi: 10.1074/mcp.M113.036442. Epub 2014 Mar 12. Mol Cell Proteomics. 2014. PMID: 24623590 Free PMC article.
-
Experimental approaches to studying translation in plant semi-autonomous organelles.J Exp Bot. 2024 Sep 11;75(17):5175-5187. doi: 10.1093/jxb/erae151. J Exp Bot. 2024. PMID: 38592734 Free PMC article. Review.
-
Light-Dependent Translation Change of Arabidopsis psbA Correlates with RNA Structure Alterations at the Translation Initiation Region.Cells. 2021 Feb 4;10(2):322. doi: 10.3390/cells10020322. Cells. 2021. PMID: 33557293 Free PMC article.
-
A PPR Protein ACM1 Is Involved in Chloroplast Gene Expression and Early Plastid Development in Arabidopsis.Int J Mol Sci. 2021 Mar 3;22(5):2512. doi: 10.3390/ijms22052512. Int J Mol Sci. 2021. PMID: 33802303 Free PMC article.
References
-
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

