Coronary heart disease is associated with a mutation in mitochondrial tRNA
- PMID: 23736300
- PMCID: PMC3781636
- DOI: 10.1093/hmg/ddt256
Coronary heart disease is associated with a mutation in mitochondrial tRNA
Abstract
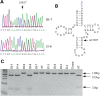
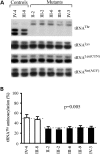
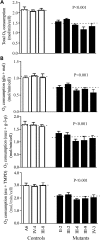
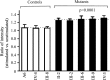
Coronary heart disease (CHD) is the leading cause of death worldwide. Mitochondrial genetic determinant for the development of CHD remains poorly explored. We report there the clinical, genetic, molecular and biochemical characterization of a four-generation Chinese family with maternally inherited CHD. Thirteen of 32 adult members in this family exhibited variable severity and age-at-onset of CHD. Mutational analysis of their mitochondrial genomes identified the tRNA(Thr) 15927G>A mutation belonging to the Eastern Asian haplogroup B5. The anticipated destabilization of a highly conserved base-pairing (28C-42G) by the 15927G>A mutation affects structure and function of tRNA(Thr). Northern analysis revealed ≈80% decrease in the steady-state level of tRNA(Thr) in the mutant cell lines carrying the 15927G>A mutation. The 15927G>A mutation changed the conformation of tRNA(Thr), as suggested by slower electrophoretic mobility of mutated tRNA with respect to the wild-type molecule. In addition, ∼39% reduction in aminoacylated efficiency of tRNA(Thr) was observed in mutant cells derived from this Chinese family. An in vivo mitochondrial protein labeling analysis showed ∼53% reduction in the rate of mitochondrial translation in mutant cells. The impaired mitochondrial protein synthesis leads to defects in overall respiratory capacity or malate/glutamate-promoted respiration or succinate/glycerol-3-phosphate-promoted respiration, or N,N,N',N'-tetramethyl-pphenylenediamine/ascorbate-promoted respiration in mutant cells. An increasing production of reactive oxygen species was observed in the mutant cells carrying the 15927G>A mutation. These results provide the direct evidence that the tRNA(Thr) 15927G>A mutation is associated with CHD. Our findings may provide new insights into pathophysiology and intervention targets of this disorder.
Figures



References
-
- Lopez A.D., Mathers C.D., Ezzati M., Jamison D.T., Murray C.J. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. - PubMed
-
- Zhang X.H., Lu Z.L., Liu L. Coronary heart disease in China. Heart. 2008;94:1126–1131. - PubMed
-
- Sing C.F., Stengård J.H., Kardia S.L. Genes, environment, and cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2003;23:1190–1196. - PubMed
-
- Wilson P.W., D'Agostino R.B., Levy D., Belanger A.M., Silbershatz H., Kannel W.B. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. - PubMed
-
- Khot U.N., Khot M.B., Bajzer C.T., Sapp S.K., Ohman E.M., Brener S.J., Ellis S.G., Lincoff A.M., Topol E.J. Prevalence of conventional risk factors in patients with coronary heart disease. J. Am. Med. Assoc. 2003;290:898–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

