Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging
- PMID: 23737254
- PMCID: PMC4212311
- DOI: 10.1002/prot.24336
Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging
Abstract
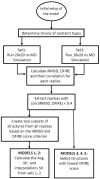
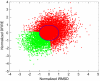
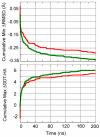
We used molecular dynamics (MD) simulations for structure refinement of Critical Assessment of Techniques for Protein Structure Prediction 10 (CASP10) targets. Refinement was achieved by selecting structures from the MD-based ensembles followed by structural averaging. The overall performance of this method in CASP10 is described, and specific aspects are analyzed in detail to provide insight into key components. In particular, the use of different restraint types, sampling from multiple short simulations versus a single long simulation, the success of a quality assessment criterion, the application of scoring versus averaging, and the impact of a final refinement step are discussed in detail.
Keywords: CASP; protein; quality assessment; scoring; structure prediction.
Copyright © 2013 Wiley Periodicals, Inc.
Figures

References
-
- Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79:37–58. - PubMed
-
- Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61:225–236. - PubMed
-
- Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. - PubMed
-
- Moult J, Fidelis K, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP) - Round 6. Proteins. 2005;61:3–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

