Genome-wide characterization of adaptation and speciation in tiger swallowtail butterflies using de novo transcriptome assemblies
- PMID: 23737327
- PMCID: PMC3698933
- DOI: 10.1093/gbe/evt090
Genome-wide characterization of adaptation and speciation in tiger swallowtail butterflies using de novo transcriptome assemblies
Abstract
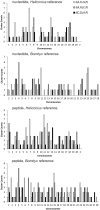
Hybrid speciation appears to be rare in animals, yet characterization of possible examples offers to shed light on the genomic consequences of this unique phenomenon, as well as more general processes such as the role of adaptation in speciation. Here, we first generate transcriptome assemblies for a putative hybrid butterfly species, Papilio appalachiensis, its parental species, P. glaucus and P. canadensis, and an outgroup, P. polytes. Then, we use these data to infer genome-wide patterns of introgression and genomic mosaicism using both phylogenetic and population genetic approaches. Our results reveal that there is little genetic divergence among all three of the focal species, but the subset of gene trees that strongly support a specific tree topology suggest widespread sharing of genetic variation between P. appalachiensis and both parental species, likely as a result of hybrid speciation. We also find evidence for substantial shared genetic variation between P. glaucus and P. canadensis, which may be due to gene flow or ancestral variation. Consistent with previous work, we show that P. applachiensis is more similar to P. canadensis at Z-linked genes and more similar to P. glaucus at mitochondrial genes. We also identify a variety of targets of adaptive evolution, which appear to be enriched for traits that are likely to be important in the evolution of this butterfly system, such as pigmentation, hormone sensitivity, developmental processes, and cuticle formation. Overall, our results provide a genome-wide portrait of divergence and introgression associated with adaptation and speciation in an iconic butterfly radiation.
Keywords: Papilio; adaptation; hybrid speciation; introgression; transcriptome.
Figures

Similar articles
-
Sex chromosome mosaicism and hybrid speciation among tiger swallowtail butterflies.PLoS Genet. 2011 Sep;7(9):e1002274. doi: 10.1371/journal.pgen.1002274. Epub 2011 Sep 8. PLoS Genet. 2011. PMID: 21931567 Free PMC article.
-
Allochronic isolation and incipient hybrid speciation in tiger swallowtail butterflies.Oecologia. 2010 Feb;162(2):523-31. doi: 10.1007/s00442-009-1493-8. Epub 2009 Nov 24. Oecologia. 2010. PMID: 19937057
-
Comparative Transcriptomics Provides Insights into Reticulate and Adaptive Evolution of a Butterfly Radiation.Genome Biol Evol. 2019 Oct 1;11(10):2963-2975. doi: 10.1093/gbe/evz202. Genome Biol Evol. 2019. PMID: 31518398 Free PMC article.
-
Review. Hybrid trait speciation and Heliconius butterflies.Philos Trans R Soc Lond B Biol Sci. 2008 Sep 27;363(1506):3047-54. doi: 10.1098/rstb.2008.0065. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 18579480 Free PMC article. Review.
-
Introgression of wing pattern alleles and speciation via homoploid hybridization in Heliconius butterflies: a review of evidence from the genome.Proc Biol Sci. 2012 Dec 12;280(1752):20122302. doi: 10.1098/rspb.2012.2302. Print 2013 Feb 7. Proc Biol Sci. 2012. PMID: 23235702 Free PMC article. Review.
Cited by
-
The complete mitochondrial genome of Lerema accius and its phylogenetic implications.PeerJ. 2016 Jan 7;4:e1546. doi: 10.7717/peerj.1546. eCollection 2016. PeerJ. 2016. PMID: 26788426 Free PMC article.
-
A cryptic new species of tiger swallowtail (Lepidoptera, Papilionidae) from eastern North America.Zookeys. 2025 Feb 14;1228:69-97. doi: 10.3897/zookeys.1228.142202. eCollection 2025. Zookeys. 2025. PMID: 39990191 Free PMC article.
-
Repeated Reticulate Evolution in North American Papilio machaon Group Swallowtail Butterflies.PLoS One. 2015 Oct 30;10(10):e0141882. doi: 10.1371/journal.pone.0141882. eCollection 2015. PLoS One. 2015. PMID: 26517268 Free PMC article.
-
Counteracting forces of introgressive hybridization and interspecific competition shape the morphological traits of cryptic Iberian Eptesicus bats.Sci Rep. 2022 Jul 8;12(1):11695. doi: 10.1038/s41598-022-15412-2. Sci Rep. 2022. PMID: 35803997 Free PMC article.
-
Comparative genomics of the mimicry switch in Papilio dardanus.Proc Biol Sci. 2014 Jul 22;281(1787):20140465. doi: 10.1098/rspb.2014.0465. Epub 2014 Jun 11. Proc Biol Sci. 2014. PMID: 24920480 Free PMC article.
References
-
- Abbott RJ, Rieseberg LH. Hybrid speciation. Encyclopaedia of Life Sciences (eLS) Chichester (UK): John Wiley & Sons; 2012.
-
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. - PubMed
-
- Brower JVZ. Experimental studies of mimicry in some North American butterflies: Part II. Battus philenor and Papilio troilus, P. polyxenes and P. glaucus. Evolution. 1958;12:123–136.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

