Cooperative working of bacterial chromosome replication proteins generated by a reconstituted protein expression system
- PMID: 23737447
- PMCID: PMC3737561
- DOI: 10.1093/nar/gkt489
Cooperative working of bacterial chromosome replication proteins generated by a reconstituted protein expression system
Abstract
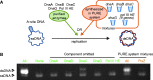
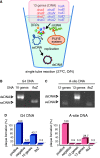
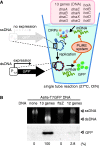
Replication of all living cells relies on the multirounds flow of the central dogma. Especially, expression of DNA replication proteins is a key step to circulate the processes of the central dogma. Here we achieved the entire sequential transcription-translation-replication process by autonomous expression of chromosomal DNA replication machineries from a reconstituted transcription-translation system (PURE system). We found that low temperature is essential to express a complex protein, DNA polymerase III, in a single tube using the PURE system. Addition of the 13 genes, encoding initiator, DNA helicase, helicase loader, RNA primase and DNA polymerase III to the PURE system gave rise to a DNA replication system by a coupling manner. An artificial genetic circuit demonstrated that the DNA produced as a result of the replication is able to provide genetic information for proteins, indicating the in vitro central dogma can sequentially undergo two rounds.
Figures

References
-
- Sethi VS, Zillig W, Bauer H. Dissociation and reconstitution of active DNA-dependent RNA-polymerase from E. coli. FEBS Lett. 1970;8:236–239. - PubMed
-
- Kaguni JM, Kornberg A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell. 1984;38:183–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

