Discovering weighted patterns in intron sequences using self-adaptive harmony search and back-propagation algorithms
- PMID: 23737711
- PMCID: PMC3662175
- DOI: 10.1155/2013/249034
Discovering weighted patterns in intron sequences using self-adaptive harmony search and back-propagation algorithms
Abstract
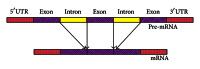
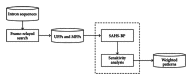
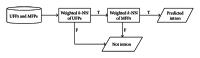
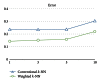
A hybrid self-adaptive harmony search and back-propagation mining system was proposed to discover weighted patterns in human intron sequences. By testing the weights under a lazy nearest neighbor classifier, the numerical results revealed the significance of these weighted patterns. Comparing these weighted patterns with the popular intron consensus model, it is clear that the discovered weighted patterns make originally the ambiguous 5SS and 3SS header patterns more specific and concrete.
Figures





References
-
- Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Current Opinion in Cell Biology. 2001;13(3):302–309. - PubMed
-
- Sharp PA. The discovery of split genes and RNA splicing. Trends in Biochemical Sciences. 2005;30(6):279–281. - PubMed
-
- Sharp PA. Splicing of messenger RNA precursors. Science. 1987;235(4790):766–771. - PubMed
-
- Zhang MQ. Statistical features of human exons and their flanking regions. Human Molecular Genetics. 1998;7(5):919–932. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

