New susceptibility locus for obesity and dyslipidaemia on chromosome 3q22.3
- PMID: 23738802
- PMCID: PMC3681549
- DOI: 10.1186/1479-7364-7-15
New susceptibility locus for obesity and dyslipidaemia on chromosome 3q22.3
Abstract
Background: The muscle Ras (MRAS) gene resides on chromosome 3q22.3 and encodes a member of the membrane-associated Ras small GTPase proteins, which function as signal transducers in multiple processes including cell growth and differentiation. Its role in cardiovascular disease is not fully understood yet. In a preliminary study in heterozygous familial hypercholesterolaemia, we identified a locus linking the early onset of coronary artery disease (CAD) to chromosome 3q.22 and elected to sequence the MRAS gene using the MegaBACE DNA analysis system. In the present study, we investigated the association of seven single-nucleotide polymorphisms (SNPs) at this locus with CAD and its dyslipidaemia-related risk traits in 4,650 Saudi angiographed individuals using TaqMan assays by the Applied Biosystems real-time Prism 7900HT Sequence Detection System.
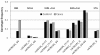
Results: Among the studied SNPs, rs6782181 (p = 0.017) and rs9818870T (p = 0.009) were associated with CAD following adjustment for sex, age and other confounding risk factors. The rs6782181_GG also conferred risk for obesity (1,764 cases vs. 2,586 controls) [1.16(1.03-1.30); p = 0.017], hypercholesterolaemia (1,686 vs. 2,744) [1.23(1.02-1.47); p = 0.019], hypertriglyceridaemia (1,155 vs. 3,496) [1.29(1.01-1.45); p = 0.043] and low high-density lipoprotein-cholesterol (lHDL-chol) levels (1,935 vs. 2,401) [1.15(1.02-1.30); p = 0.023] after adjustment. Additionally, rs253662_(CT+TT) [1.16(1.01-1.32); p = 0.030] was associated with lHDL-chol levels. Interestingly, rs253662 (p = 0.014) and rs6782181 (p = 0.019) were protective against acquiring high low-density lipoprotein-cholesterol (hLDL-chol) levels (p = 0.014), while rs1720819 showed similar effects against CAD (p < 0.0001). More importantly, a 7-mer haplotype, ACCTGAC (χ2 = 7.66; p = 0.0056), constructed from the studied SNPs, its 6-mer derivative CCTGAC (χ2 = 6.90; p = 0.0086) and several other shorter derivatives conferred risk for obesity. hLDL-chol was weakly linked to CTAA (χ2 = 3.79; p = 0.052) and CCT (χ2 = 4.32; p = 0.038), while several other haplotypes were protective against both obesity and hLDL-chol level.
Conclusion: Our results demonstrate that the genomic locus for the MRAS gene confers risk for CAD, obesity and dyslipidaemia and point to the possible involvement of other genes or regulatory elements at this locus, rather than changes in the M-Ras protein function, in these events.
Figures

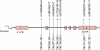

References
-
- Quilliam LA, Castro AF, Rogers-Graham KS, Martin CB, Der CJ, Bi C. M-Ras/R-Ras3, a transforming ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J Biol Chem. 1999;274(34):23850–23857. doi: 10.1074/jbc.274.34.23850. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

