Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice
- PMID: 23740943
- PMCID: PMC3781640
- DOI: 10.1093/hmg/ddt261
Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice
Abstract
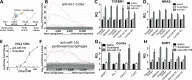
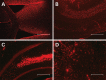
microRNAs (miRNAs) are dysregulated in a variety of disease states, suggesting that this newly discovered class of gene expression repressors may be viable therapeutic targets. A microarray of miRNA changes in ALS-model superoxide dismutase 1 (SOD1)(G93A) rodents identified 12 miRNAs as significantly changed. Six miRNAs tested in human ALS tissues were confirmed increased. Specifically, miR-155 was increased 5-fold in mice and 2-fold in human spinal cords. To test miRNA inhibition in the central nervous system (CNS) as a potential novel therapeutic, we developed oligonucleotide-based miRNA inhibitors (anti-miRs) that could inhibit miRNAs throughout the CNS and in the periphery. Anti-miR-155 caused global derepression of targets in peritoneal macrophages and, following intraventricular delivery, demonstrated widespread functional distribution in the brain and spinal cord. After treating SOD1(G93A) mice with anti-miR-155, we significantly extended survival by 10 days and disease duration by 15 days (38%) while a scrambled control anti-miR did not significantly improve survival or disease duration. Therefore, antisense oligonucleotides may be used to successfully inhibit miRNAs throughout the brain and spinal cord, and miR-155 is a promising new therapeutic target for human ALS.
Figures



References
-
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Rothstein J.D. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann. Neurol. 2009;65(Suppl 1):S3–S9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

