Toward eliminating HLA class I expression to generate universal cells from allogeneic donors
- PMID: 23741009
- PMCID: PMC3750336
- DOI: 10.1182/blood-2013-03-478255
Toward eliminating HLA class I expression to generate universal cells from allogeneic donors
Abstract
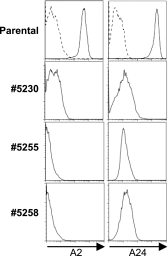
Long-term engraftment of allogeneic cells necessitates eluding immune-mediated rejection, which is currently achieved by matching for human leukocyte antigen (HLA) expression, immunosuppression, and/or delivery of donor-derived cells to sanctuary sites. Genetic engineering provides an alternative approach to avoid clearance of cells that are recognized as "non-self" by the recipient. To this end, we developed designer zinc finger nucleases and employed a "hit-and-run" approach to genetic editing for selective elimination of HLA expression. Electro-transfer of mRNA species coding for these engineered nucleases completely disrupted expression of HLA-A on human T cells, including CD19-specific T cells. The HLA-A(neg) T-cell pools can be enriched and evade lysis by HLA-restricted cytotoxic T-cell clones. Recognition by natural killer cells of cells that had lost HLA expression was circumvented by enforced expression of nonclassical HLA molecules. Furthermore, we demonstrate that zinc finger nucleases can eliminate HLA-A expression from embryonic stem cells, which broadens the applicability of this strategy beyond infusing HLA-disparate immune cells. These findings establish that clinically appealing cell types derived from donors with disparate HLA expression can be genetically edited to evade an immune response and provide a foundation whereby cells from a single donor can be administered to multiple recipients.
Figures





References
-
- Gaspar HB, Cooray S, Gilmour KC, et al. Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci Transl Med. 2011;3(97):97ra80. - PubMed
-
- Rama P, Matuska S, Paganoni G, Spinelli A, De Luca M, Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med. 2010;363(2):147–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA16672/CA/NCI NIH HHS/United States
- CA120956/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- R37 CA084198/CA/NCI NIH HHS/United States
- R01 CA120956/CA/NCI NIH HHS/United States
- R01 CA141303/CA/NCI NIH HHS/United States
- CA124782/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA124782/CA/NCI NIH HHS/United States
- R01 HD045022/HD/NICHD NIH HHS/United States
- R01 CA084198/CA/NCI NIH HHS/United States
- R01CA087869/CA/NCI NIH HHS/United States
- P01 CA148600/CA/NCI NIH HHS/United States
- R01 CA087869/CA/NCI NIH HHS/United States
- R21 CA116127/CA/NCI NIH HHS/United States
- R33 CA116127/CA/NCI NIH HHS/United States
- CA141303/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

