Identification of novel tissue-specific genes by analysis of microarray databases: a human and mouse model
- PMID: 23741331
- PMCID: PMC3669334
- DOI: 10.1371/journal.pone.0064483
Identification of novel tissue-specific genes by analysis of microarray databases: a human and mouse model
Abstract
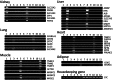
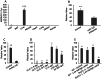
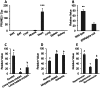
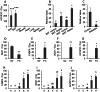
Understanding the tissue-specific pattern of gene expression is critical in elucidating the molecular mechanisms of tissue development, gene function, and transcriptional regulations of biological processes. Although tissue-specific gene expression information is available in several databases, follow-up strategies to integrate and use these data are limited. The objective of the current study was to identify and evaluate novel tissue-specific genes in human and mouse tissues by performing comparative microarray database analysis and semi-quantitative PCR analysis. We developed a powerful approach to predict tissue-specific genes by analyzing existing microarray data from the NCBI's Gene Expression Omnibus (GEO) public repository. We investigated and confirmed tissue-specific gene expression in the human and mouse kidney, liver, lung, heart, muscle, and adipose tissue. Applying our novel comparative microarray approach, we confirmed 10 kidney, 11 liver, 11 lung, 11 heart, 8 muscle, and 8 adipose specific genes. The accuracy of this approach was further verified by employing semi-quantitative PCR reaction and by searching for gene function information in existing publications. Three novel tissue-specific genes were discovered by this approach including AMDHD1 (amidohydrolase domain containing 1) in the liver, PRUNE2 (prune homolog 2) in the heart, and ACVR1C (activin A receptor, type IC) in adipose tissue. We further confirmed the tissue-specific expression of these 3 novel genes by real-time PCR. Among them, ACVR1C is adipose tissue-specific and adipocyte-specific in adipose tissue, and can be used as an adipocyte developmental marker. From GEO profiles, we predicted the processes in which AMDHD1 and PRUNE2 may participate. Our approach provides a novel way to identify new sets of tissue-specific genes and to predict functions in which they may be involved.
Conflict of interest statement
Figures

Similar articles
-
Identification of CTLA2A, DEFB29, WFDC15B, SERPINA1F and MUP19 as Novel Tissue-Specific Secretory Factors in Mouse.PLoS One. 2015 May 6;10(5):e0124962. doi: 10.1371/journal.pone.0124962. eCollection 2015. PLoS One. 2015. PMID: 25946105 Free PMC article.
-
MGFM: a novel tool for detection of tissue and cell specific marker genes from microarray gene expression data.BMC Genomics. 2015 Aug 28;16(1):645. doi: 10.1186/s12864-015-1785-9. BMC Genomics. 2015. PMID: 26314578 Free PMC article.
-
Differential expression of cyclin G2, cyclin-dependent kinase inhibitor 2C and peripheral myelin protein 22 genes during adipogenesis.Animal. 2014 May;8(5):800-9. doi: 10.1017/S1751731114000469. Animal. 2014. PMID: 24739352
-
Genetic dissection of gene regulation in multiple mouse tissues.Mamm Genome. 2006 Jun;17(6):490-5. doi: 10.1007/s00335-005-0186-9. Epub 2006 Jun 12. Mamm Genome. 2006. PMID: 16783630 Review.
-
Databases of free expression.Mamm Genome. 2006 Dec;17(12):1141-6. doi: 10.1007/s00335-006-0043-5. Epub 2006 Dec 1. Mamm Genome. 2006. PMID: 17143588 Review.
Cited by
-
The Utility of Resolving Asthma Molecular Signatures Using Tissue-Specific Transcriptome Data.G3 (Bethesda). 2020 Nov 5;10(11):4049-4062. doi: 10.1534/g3.120.401718. G3 (Bethesda). 2020. PMID: 32900903 Free PMC article.
-
Analysis of Effect of Schisandra in the Treatment of Myocardial Infarction Based on Three-Mode Gene Ontology Network.Front Pharmacol. 2019 Mar 20;10:232. doi: 10.3389/fphar.2019.00232. eCollection 2019. Front Pharmacol. 2019. PMID: 30949047 Free PMC article.
-
Screening and Identification of Muscle-Specific Candidate Genes via Mouse Microarray Data Analysis.Front Vet Sci. 2021 Dec 13;8:794628. doi: 10.3389/fvets.2021.794628. eCollection 2021. Front Vet Sci. 2021. PMID: 34966817 Free PMC article.
-
Genome-wide association study of genetic variants related to anthracycline-induced cardiotoxicity in early breast cancer.Cancer Sci. 2020 Jul;111(7):2579-2587. doi: 10.1111/cas.14446. Epub 2020 Jun 20. Cancer Sci. 2020. PMID: 32378780 Free PMC article.
-
Robust and rigorous identification of tissue-specific genes by statistically extending tau score.BioData Min. 2022 Dec 9;15(1):31. doi: 10.1186/s13040-022-00315-9. BioData Min. 2022. PMID: 36494766 Free PMC article.
References
-
- Niehrs C, Pollet N (1999) Synexpression groups in eukaryotes. Nature 402: 483–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

