Latent transforming growth factor β-binding protein 4 is downregulated in esophageal cancer via promoter methylation
- PMID: 23741501
- PMCID: PMC3669142
- DOI: 10.1371/journal.pone.0065614
Latent transforming growth factor β-binding protein 4 is downregulated in esophageal cancer via promoter methylation
Abstract
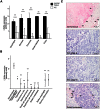
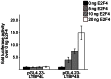
Latent transforming growth factor β-binding protein 4 (LTBP4) is an extracellular matrix molecule that is a member of important connective tissue networks and is needed for the correct folding and the secretion of TGF-β1. LTBP4 is downregulated in carcinomas of various tissues. Here we show that LTBP4 is also downregulated in adenocarcinomas and squamous cell carcinomas of the esophagus in vitro and in vivo. Re-expression of LTBP4 in esophageal cancer cell lines reduced cell migration ability, whereas cell viability and cell proliferation remained unchanged. Hypermethylation of the promoter regions of the two main human LTBP4 transcriptional forms, LTBP4L and LTBP4S, was found to be involved in LTBP4 silencing. Detailed investigations of the methylation patterns of the promoter regions of LTBP4L and LTBP4S identified GATA1, SP1, E2F4 and SMAD3 as potential transcription factors involved in LTBP4 expression. In in vitro transcription factor activity studies we discovered E2F4 as novel powerful regulator for LTBP4S expression.
Conflict of interest statement
Figures




References
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. - PubMed
-
- Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, et al. (2012) Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 62: 220–241. - PubMed
-
- Gavin AT, Francisci S, Foschi R, Donnelly DW, Lemmens V, et al... (2012) Oesophageal cancer survival in Europe: A EUROCARE-4 study. Cancer Epidemiol. - PubMed
-
- Holmes RS, Vaughan TL (2007) Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol 17: 2–9. - PubMed
-
- Groblewska M, Siewko M, Mroczko B, Szmitkowski M (2012) The role of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia Histochem Cytobiol 50: 12–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

