The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine
- PMID: 23741619
- PMCID: PMC3667578
- DOI: 10.7554/eLife.00731
The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine
Erratum in
- Elife. 2013;2:e01108
Abstract
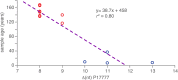
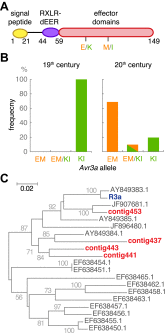
Phytophthora infestans, the cause of potato late blight, is infamous for having triggered the Irish Great Famine in the 1840s. Until the late 1970s, P. infestans diversity outside of its Mexican center of origin was low, and one scenario held that a single strain, US-1, had dominated the global population for 150 years; this was later challenged based on DNA analysis of historical herbarium specimens. We have compared the genomes of 11 herbarium and 15 modern strains. We conclude that the 19th century epidemic was caused by a unique genotype, HERB-1, that persisted for over 50 years. HERB-1 is distinct from all examined modern strains, but it is a close relative of US-1, which replaced it outside of Mexico in the 20th century. We propose that HERB-1 and US-1 emerged from a metapopulation that was established in the early 1800s outside of the species' center of diversity. DOI:http://dx.doi.org/10.7554/eLife.00731.001.
Keywords: Herbarium; Other; Phytophthora infestans; Solanum tuberosum; ancient DNA.
Conflict of interest statement
DW: Deputy editor,
The other authors declare that no competing interests exist.
Figures

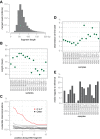
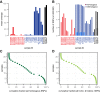

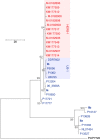
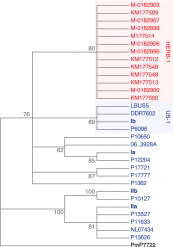





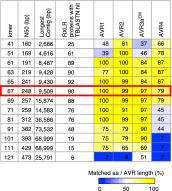
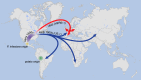
Comment in
-
The early days of late blight.Elife. 2013 Jun 18;2:e00954. doi: 10.7554/eLife.00954. Elife. 2013. PMID: 23795302 Free PMC article.
References
-
- Bourke PMA. 1964. Emergence of potato blight, 1843-46. Nature 203:805–8. 10.1038/203805a0 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

